Immunogenic neoantigens derived from gene fusions stimulate T cell responses
- PMID: 31011208
- PMCID: PMC6558662
- DOI: 10.1038/s41591-019-0434-2
Immunogenic neoantigens derived from gene fusions stimulate T cell responses
Abstract
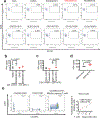
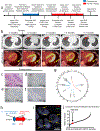
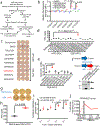
Anti-tumor immunity is driven by self versus non-self discrimination. Many immunotherapeutic approaches to cancer have taken advantage of tumor neoantigens derived from somatic mutations. Here, we demonstrate that gene fusions are a source of immunogenic neoantigens that can mediate responses to immunotherapy. We identified an exceptional responder with metastatic head and neck cancer who experienced a complete response to immune checkpoint inhibitor therapy, despite a low mutational load and minimal pre-treatment immune infiltration in the tumor. Using whole-genome sequencing and RNA sequencing, we identified a novel gene fusion and demonstrated that it produces a neoantigen that can specifically elicit a host cytotoxic T cell response. In a cohort of head and neck tumors with low mutation burden, minimal immune infiltration and prevalent gene fusions, we also identified gene fusion-derived neoantigens that generate cytotoxic T cell responses. Finally, analyzing additional datasets of fusion-positive cancers, including checkpoint-inhibitor-treated tumors, we found evidence of immune surveillance resulting in negative selective pressure against gene fusion-derived neoantigens. These findings highlight an important class of tumor-specific antigens and have implications for targeting gene fusion events in cancers that would otherwise be less poised for response to immunotherapy, including cancers with low mutational load and minimal immune infiltration.
Conflict of interest statement
Competing interests:
K.-W.L. and J.S.S. are now full-time employees of Regeneron Pharmaceuticals. R.M.Srivastava received speaker fees and travel reimbursement from Innovent Biologics, Inc. A.L.H. receives research funding from Eisai, Bristol-Myers Squibb, Kura Oncology, AstraZeneca, Genentech Roche, Celldex, Pfizer, Lilly and Bayer; consulting fees from Bristol-Myers Squibb, Merck, Novartis, AstraZeneca, Regeneron, Sanofi Aventis, Sun Pharmaceuticals, Eisai, Genentech/Roche, Genzyme and Ayala Pharmaceuticals; and travel fees from Ignyta and Kura Oncology. J.J.H’s spouse is a full-time employee of Regeneron Pharmaceuticals. R.M.Samstein, T.A.C. and L.G.T.M. are inventors on a provisional patent application (62/569,053) filed by Memorial Sloan Kettering (MSK) relating to the use of TMB in cancer immunotherapy. D.C. and T.A.C. are inventors on a PCT patent application (PCT/US2015/062208) filed by MSK relating to the use of TMB in cancer immunotherapy. MSK and the inventors may receive a share of commercialization revenue from license agreements relating to these patent applications. TAC is a co-founder of Gritstone Oncology and holds equity. TAC acknowledges grant funding from Bristol-Myers Squibb, AstraZeneca, Illumina, Pfizer, An2H, and Eisai. TAC has served as an advisor for Bristol-Myers Squibb, Illumina, Eisai, and An2H. L.G.T.M. received consulting fees from Rakuten Aspyrian and speaker fees from Physician Educational Resources.
Figures











Comment in
-
Gene-fusion neoantigens stimulate T cells.Nat Rev Cancer. 2019 Jul;19(7):364. doi: 10.1038/s41568-019-0160-6. Nat Rev Cancer. 2019. PMID: 31160733 No abstract available.
References
Methods-only References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

