Immunoinformatics Approach for Multiepitopes Vaccine Prediction against Glycoprotein B of Avian Infectious Laryngotracheitis Virus
- PMID: 31011331
- PMCID: PMC6442309
- DOI: 10.1155/2019/1270485
Immunoinformatics Approach for Multiepitopes Vaccine Prediction against Glycoprotein B of Avian Infectious Laryngotracheitis Virus
Abstract
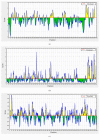
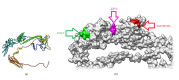
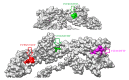
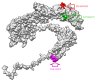
Infectious laryngotracheitis virus (ILTV) is a gallid herpesvirus type 1, a member of the genus Iltovirus. It causes an infection in the upper respiratory tract mainly trachea which results in significant economic losses in the poultry industry worldwide. Vaccination against ILTV produced latent infected carriers' birds, which become a source of virus transmission to nonvaccinated flocks. Thus this study aimed to design safe multiepitopes vaccine against glycoprotein B of ILT virus using immunoinformatic tools. Forty-four sequences of complete envelope glycoprotein B were retrieved from GenBank of National Center for Biotechnology Information (NCBI) and aligned for conservancy by multiple sequence alignment (MSA). Immune Epitope Database (IEDB) analysis resources were used to predict and analyze candidate epitopes that could act as a promising peptide vaccine. For B cell epitopes, thirty-one linear epitopes were predicted using Bepipred. However eight epitopes were found to be on both surface and antigenic epitopes using Emini surface accessibility and antigenicity, respectively. Three epitopes ( 190 KKLP 193 , 386 YSSTHVRS 393 , and 317 KESV 320 ) were proposed as B cell epitopes. For T cells several epitopes were interacted with MHC class I with high affinity and specificity, but the best recognized epitopes were 118 YVFNVTLYY 126 , 335 VSYKNSYHF 343 , and 622 YLLYEDYTF 630 . MHC-II binding epitopes, 301 FLTDEQFTI 309 , 277 FLEIANYQV 285 , and 743 IASFLSNPF 751 , were proposed as promising epitopes due to their high affinity for MHC-II molecules. Moreover the docked ligand epitopes from MHC-1 molecule exhibited high binding affinity with the receptors; BF chicken alleles (BF2 2101 and 0401) expressed by the lower global energy of the molecules. In this study nine epitopes were predicted as promising vaccine candidate against ILTV. In vivo and in vitro studies are required to support the effectiveness of these predicted epitopes as a multipeptide vaccine through clinical trials.
Figures




Similar articles
-
Vaccine design of coronavirus spike (S) glycoprotein in chicken: immunoinformatics and computational approaches.Transl Med Commun. 2020;5(1):13. doi: 10.1186/s41231-020-00063-0. Epub 2020 Aug 27. Transl Med Commun. 2020. PMID: 32869000 Free PMC article.
-
A recombinant Newcastle disease virus (NDV) expressing infectious laryngotracheitis virus (ILTV) surface glycoprotein D protects against highly virulent ILTV and NDV challenges in chickens.Vaccine. 2014 Jun 12;32(28):3555-63. doi: 10.1016/j.vaccine.2014.04.068. Epub 2014 Apr 30. Vaccine. 2014. PMID: 24793943
-
Prediction of promiscuous T-cell epitopes in the Zika virus polyprotein: An in silico approach.Asian Pac J Trop Med. 2016 Sep;9(9):844-850. doi: 10.1016/j.apjtm.2016.07.004. Epub 2016 Jul 26. Asian Pac J Trop Med. 2016. PMID: 27633296
-
Immune responses to infectious laryngotracheitis virus.Dev Comp Immunol. 2013 Nov;41(3):454-62. doi: 10.1016/j.dci.2013.03.022. Epub 2013 Apr 6. Dev Comp Immunol. 2013. PMID: 23567343 Review.
-
Avian infectious laryngotracheitis.Rev Sci Tech. 2000 Aug;19(2):483-92. doi: 10.20506/rst.19.2.1229. Rev Sci Tech. 2000. PMID: 10935275 Review.
Cited by
-
Design of a Multi-Epitope Vaccine Candidate Against Infectious Laryngotracheitis Virus Affecting Poultry by Computational Approaches.Biology (Basel). 2025 Jun 25;14(7):765. doi: 10.3390/biology14070765. Biology (Basel). 2025. PMID: 40723326 Free PMC article.
-
Recombinase-aided amplification-lateral flow dipstick assay-a specific and sensitive method for visual detection of avian infectious laryngotracheitis virus.Poult Sci. 2021 Mar;100(3):100895. doi: 10.1016/j.psj.2020.12.008. Epub 2020 Dec 9. Poult Sci. 2021. PMID: 33518305 Free PMC article.
-
Execution and Design of an Anti HPIV-1 Vaccine with Multiple Epitopes Triggering Innate and Adaptive Immune Responses: An Immunoinformatic Approach.Vaccines (Basel). 2022 May 29;10(6):869. doi: 10.3390/vaccines10060869. Vaccines (Basel). 2022. PMID: 35746477 Free PMC article.
-
A Computational Reverse Vaccinology Approach for the Design and Development of Multi-Epitopic Vaccine Against Avian Pathogen Mycoplasma gallisepticum.Front Vet Sci. 2021 Oct 26;8:721061. doi: 10.3389/fvets.2021.721061. eCollection 2021. Front Vet Sci. 2021. PMID: 34765664 Free PMC article.
-
Vaccine design of coronavirus spike (S) glycoprotein in chicken: immunoinformatics and computational approaches.Transl Med Commun. 2020;5(1):13. doi: 10.1186/s41231-020-00063-0. Epub 2020 Aug 27. Transl Med Commun. 2020. PMID: 32869000 Free PMC article.
References
-
- Hidalgo H. Infectious laryngotracheitis: a review. Revista Brasileira de Ciência Avícola. 2003;5(3):157–168. doi: 10.1590/S1516-635X2003000300001. - DOI
-
- Oldoni I., Rodríguez-Avila A., Riblet S., García M. Characterization of infectious laryngotracheitis virus (ILTV) isolates from commercial poultry by polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) Avian Diseases. 2008;52(1):59–63. doi: 10.1637/8054-070607-Reg. - DOI - PubMed
-
- Parra S., Nuñez L., Ferreira A. Epidemiology of avian infectious laryngotracheitis with special focus to south america: an update. Revista Brasileira de Ciência Avícola. 2016;18(4):551–562. doi: 10.1590/1806-9061-2016-0224. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
