Circular RNA cMras inhibits lung adenocarcinoma progression via modulating miR-567/PTPRG regulatory pathway
- PMID: 31012177
- PMCID: PMC6536402
- DOI: 10.1111/cpr.12610
Circular RNA cMras inhibits lung adenocarcinoma progression via modulating miR-567/PTPRG regulatory pathway
Abstract
Objectives: Circular RNA, a type of RNA formed by a covalently closed loop, possesses sophisticated abilities of gene regulation in tumorigenesis and metastasis. However, the role of circRNAs on lung adenocarcinoma (LUAD) remains largely unknown.
Materials and methods: The role of cMras was examined both in vitro and in vivo. cMras expression in LUAD tissues was determined by quantitative real-time PCR (qRT-PCR). Downstream targets of cMras were predicted by bioinformatics tools and confirmed by RNA immunoprecipitation assay and luciferase assay. qRT-PCR and western blot assay were used to detect the expression of specific targets.
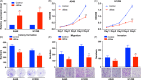
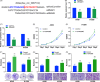
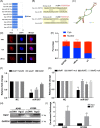
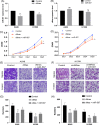
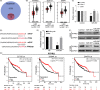
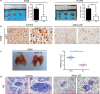
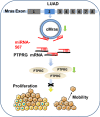
Results: Thirty-six paired LUAD and healthy tissues were collected and cMras resulted significantly downregulated in cancerous tissues. Its expression was negatively associated with tumour stages. cMras overexpression suppressed LUAD growth and metastasis, while endogenous cMras silencing resulted in the opposite effects. Bioinformatics analysis and experimental evidence confirmed that cMras was a sponge of miRNA-567 and released its direct target, PTPRG. cMras overexpression decreased miR-567 expression and subsequently increased PTPRG expression, while increased miRNA-567 expression blocked the effects induced by cMras. Moreover, PTPRG was downregulated in LUAD and patients with low PTPRG expression exhibited significantly poor prognosis. These results suggested that cMras/miR-567/PTPRG regulatory pathway might be associated to LUAD tumorigenesis and development.
Conclusions: A novel circular RNA cMras and its functions were identified, discovering a cMras/miR-567/PTPRG regulatory pathway in LUAD tumorigenesis and development.
Keywords: PTPRG; cMras; circular RNA; lung cancer; miR-567.
© 2019 The Authors. Cell Proliferation Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1‐19. - PubMed
-
- Rothschild SI. [Advanced and metastatic lung cancer – what is new in the diagnosis and therapy?]. Praxis (Bern 1994). 2015;104:745‐750. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7‐30. - PubMed
-
- Yang JX, Rastetter RH, Wilhelm D. Non‐coding RNAs: an introduction. Adv Exp Med Biol. 2016;886:13‐32. - PubMed
-
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333‐338. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

