LncRNA TRERNA1 facilitates hepatocellular carcinoma metastasis by dimethylating H3K9 in the CDH1 promoter region via the recruitment of the EHMT2/SNAI1 complex
- PMID: 31012192
- PMCID: PMC6668973
- DOI: 10.1111/cpr.12621
LncRNA TRERNA1 facilitates hepatocellular carcinoma metastasis by dimethylating H3K9 in the CDH1 promoter region via the recruitment of the EHMT2/SNAI1 complex
Abstract
Objectives: Long non-coding RNAs (LncRNAs) play an important role in hepatocellular carcinoma development, however, as a crucial driver of hepatocellular carcinoma (HCC) metastasis, their functions in tumour metastasis remain largely unknown.
Materials and methods: The lncRNA TRERNA1 expression levels were detected in HCC by quantitative real-time PCR (qPCR). The function of TRERNA1 was examined by wound-healing assays, transwell assays and tail vein injection experiments. The potential regulatory mechanisms of TRERNA1 on its target genes were explored by ChIP, RIP, IP and WB assays.
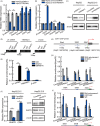
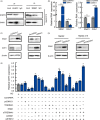
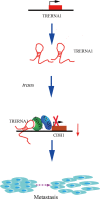
Results: Elevated TRERNA1 levels promoted HCC cell migration and invasion in vitro and in vivo. TRERNA1 recruited EHMT2 to dimethylate H3K9 in the CDH1 promoter region. Furthermore, EHMT2 bound to SNAI1 to suppress CDH1 expression in HCC cells. After inhibiting TRERNA1, the expression level of CDH1 was restored and was involved in the regulation of the EHMT2/SNAI1 complex. The level of TRERNA1 was positively correlated with tumour metastasis and was negatively correlated with the expression of CDH1 in HCC tissues.
Conclusions: For the first time, the current study reveals that TRERNA1 promotes cell metastasis and the invasion of HCC via the recruitment of EHMT2 and/or the EHMT2/SNAI1 complex to suppress CDH1. These data identify a novel mechanism that regulates TRERNA1 in metastatic HCC and provides a potential targeted therapy for HCC patients.
Keywords: EHMT2; TRERNA1; hepatocellular carcinoma; metastasis.
© 2019 The Authors. Cell Proliferation Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Consent for publication: Consent to publish has been obtained from all authors.
Figures




Similar articles
-
Long noncoding RNA metastasis-associated lung adenocarcinoma transcript 1 cooperates with enhancer of zeste homolog 2 to promote hepatocellular carcinoma development by modulating the microRNA-22/Snail family transcriptional repressor 1 axis.Cancer Sci. 2020 May;111(5):1582-1595. doi: 10.1111/cas.14372. Epub 2020 Apr 30. Cancer Sci. 2020. PMID: 32129914 Free PMC article.
-
Long non-coding RNA NRON is downregulated in HCC and suppresses tumour cell proliferation and metastasis.Biomed Pharmacother. 2018 Aug;104:102-109. doi: 10.1016/j.biopha.2018.05.006. Epub 2018 May 15. Biomed Pharmacother. 2018. PMID: 29772429
-
Long Noncoding RNA AK021443 Promotes Cell Proliferation and Migration by Regulating Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Cells.DNA Cell Biol. 2018 May;37(5):481-490. doi: 10.1089/dna.2017.4030. Epub 2018 Mar 14. DNA Cell Biol. 2018. PMID: 29638164
-
LncRNA influence sequential steps of hepatocellular carcinoma metastasis.Biomed Pharmacother. 2021 Apr;136:111224. doi: 10.1016/j.biopha.2021.111224. Epub 2021 Jan 12. Biomed Pharmacother. 2021. PMID: 33450489 Review.
-
Long noncoding RNA network: Novel insight into hepatocellular carcinoma metastasis (Review).Int J Mol Med. 2021 Jul;48(1):134. doi: 10.3892/ijmm.2021.4967. Epub 2021 May 20. Int J Mol Med. 2021. PMID: 34013360 Free PMC article. Review.
Cited by
-
Translation regulatory long non-coding RNA 1 (TRERNA1) sponges microRNA-23a to suppress granulosa cell apoptosis in premature ovarian failure.Bioengineered. 2022 Feb;13(2):2173-2180. doi: 10.1080/21655979.2021.2023802. Bioengineered. 2022. PMID: 35034562 Free PMC article.
-
The role of histone methylation in the development of digestive cancers: a potential direction for cancer management.Signal Transduct Target Ther. 2020 Aug 3;5(1):143. doi: 10.1038/s41392-020-00252-1. Signal Transduct Target Ther. 2020. PMID: 32747629 Free PMC article. Review.
-
Long Non-Coding RNAs in Liver Cancer and Nonalcoholic Steatohepatitis.Noncoding RNA. 2020 Aug 29;6(3):34. doi: 10.3390/ncrna6030034. Noncoding RNA. 2020. PMID: 32872482 Free PMC article. Review.
-
Overexpression of FOXD2-AS1 enhances proliferation and impairs differentiation of glioma stem cells by activating the NOTCH pathway via TAF-1.J Cell Mol Med. 2022 May;26(9):2620-2632. doi: 10.1111/jcmm.17268. Epub 2022 Apr 14. J Cell Mol Med. 2022. PMID: 35419917 Free PMC article.
-
Identification of differentially expressed genes and biological pathways in para-carcinoma tissues of HCC with different metastatic potentials.Oncol Lett. 2020 Jun;19(6):3799-3814. doi: 10.3892/ol.2020.11493. Epub 2020 Mar 29. Oncol Lett. 2020. PMID: 32382332 Free PMC article.
References
-
- Rahbari NN, Mehrabi A, Mollberg NM, et al. Hepatocellular carcinoma: current management and perspectives for the future. Ann Surg. 2011;253:453‐469. - PubMed
-
- El‐Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118‐1127. - PubMed
-
- Yang H, Fang F, Chang R, Yang L. MicroRNA‐140‐5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205‐217. - PubMed
-
- Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99‐111. - PubMed
MeSH terms
Substances
Grants and funding
- KYCX17_0048/the Fundamental Research Funds for the Central Universities, Postgraduate Research&Practice Innovation Program of Jiangsu Province
- KYCX18_0058/the Fundamental Research Funds for the Central Universities, Postgraduate Research&Practice Innovation Program of Jiangsu Province
- 81672414/National Natural Science Foundation of China
- 81702906/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

