Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders
- PMID: 31013090
- PMCID: PMC7444661
- DOI: 10.1021/acs.jmedchem.9b00193
Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders
Abstract
Novel pyridine- and pyrimidine-based allosteric inhibitors are reported that achieve PDE4D subtype selectivity through recognition of a single amino acid difference on a key regulatory domain, known as UCR2, that opens and closes over the catalytic site for cAMP hydrolysis. The design and optimization of lead compounds was based on iterative analysis of X-ray crystal structures combined with metabolite identification. Selectivity for the activated, dimeric form of PDE4D provided potent memory enhancing effects in a mouse model of novel object recognition with improved tolerability and reduced vascular toxicity over earlier PDE4 inhibitors that lack subtype selectivity. The lead compound, 28 (BPN14770), has entered midstage, human phase 2 clinical trials for the treatment of Fragile X Syndrome.
Figures
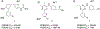
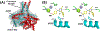


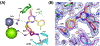
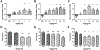
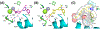

References
-
- Houslay MD, Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem Sci 2010, 35 (2), 91–100. - PubMed
-
- Gervasi N; Tchenio P; Preat T, PKA dynamics in a Drosophila learning center: coincidence detection by rutabaga adenylyl cyclase and spatial regulation by dunce phosphodiesterase. Neuron 2010, 65(4), 516–529. - PubMed
-
- Bolger G; Michaeli T; Martins T; St John T; Steiner B; Rodgers L; Riggs M; Wigler M; Ferguson K, A family of human phosphodiesterases homologous to the dunce learning and memory gene product of Drosophila melanogaster are potential targets for antidepressant drugs. Mol Cell Biol 1993, 13 (10), 6558–6571. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Medical
Molecular Biology Databases

