Involvement of p38 Activation and Mitochondria in Death of Human Leukemia Cells Induced by an Agonistic Human Monoclonal Antibody Fab Specific to TRAIL Receptor 1
- PMID: 31013630
- PMCID: PMC6515105
- DOI: 10.3390/ijms20081967
Involvement of p38 Activation and Mitochondria in Death of Human Leukemia Cells Induced by an Agonistic Human Monoclonal Antibody Fab Specific to TRAIL Receptor 1
Abstract
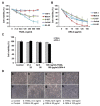
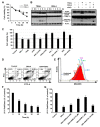
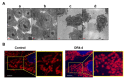
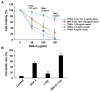
The tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces cancer cell death with minimal damage to normal cells; however, some cancer cells are resistant to TRAIL. TRAIL resistance may be overcome by agonistic antibodies to TRAIL receptors. In this study, we report the toxic effects of a novel recombinant agonistic human anti-TRAIL receptor 1 (DR4) monoclonal antibody Fab fragment, DR4-4, on various TRAIL-resistant and -sensitive cancer cell lines. The mechanisms of DR4-4 Fab-induced cell death in a human T cell leukemia cell line (Jurkat) were investigated using cell viability testing, immunoblotting, immunoassays, flow cytometry, and morphological observation. DR4-4 Fab-induced caspase-independent necrosis was observed to occur in Jurkat cells in association with p38 mitogen-activated protein kinase activation, cellular FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein degradation, decreased mitochondrial membrane potential, and increased mitochondrial reactive oxygen species production. Increased cytotoxic effects of DR4-4 Fab were observed in combination with TRAIL or γ-irradiation. Our results indicate that the novel DR4-4 Fab might overcome TRAIL-resistance and induce death in leukemia cells via cellular mechanisms different from those activated by TRAIL. DR4-4 Fab may have application as a potential therapeutic antibody fragment in single or combination therapy for cancer.
Keywords: DR4; Fab; TRAIL; TRAIL receptor 1; TRAIL-resistance; agonistic human antibody; cancer cells; mitochondria; p38.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Ewing's sarcoma family tumors are sensitive to tumor necrosis factor-related apoptosis-inducing ligand and express death receptor 4 and death receptor 5.Cancer Res. 2001 Mar 15;61(6):2704-12. Cancer Res. 2001. PMID: 11289151
-
Synergistic induction of tumor cell apoptosis by death receptor antibody and chemotherapy agent through JNK/p38 and mitochondrial death pathway.Oncogene. 2003 Apr 3;22(13):2034-44. doi: 10.1038/sj.onc.1206290. Oncogene. 2003. PMID: 12673208
-
Effect of a novel fully human monovalent antigen-binding fragment on the survival of cancer cell lines.Oncol Rep. 2007 Aug;18(2):513-7. Oncol Rep. 2007. PMID: 17611678
-
Mechanisms of resistance to TRAIL-induced apoptosis in cancer.Cancer Gene Ther. 2005 Mar;12(3):228-37. doi: 10.1038/sj.cgt.7700792. Cancer Gene Ther. 2005. PMID: 15550937 Review.
-
TRAIL receptor signalling and modulation: Are we on the right TRAIL?Cancer Treat Rev. 2009 May;35(3):280-8. doi: 10.1016/j.ctrv.2008.11.006. Epub 2008 Dec 30. Cancer Treat Rev. 2009. PMID: 19117685 Review.
References
-
- Azijli K., Yuvaraj S., van Roosmalen I., Flach K., Giovannetti E., Peters G., de Jong S., Kruyt F. MAPK p38 and JNK have opposing activities on TRAIL-induced apoptosis activation in NSCLC H460 cells that involves RIP1 and caspase-8 and is mediated by Mcl-1. Apoptosis. 2013;18:851–860. doi: 10.1007/s10495-013-0829-3. - DOI - PubMed
-
- Griffith T.S., Rauch C.T., Smolak P.J., Waugh J.Y., Boiani N., Lynch D.H., Smith C.A., Goodwin R.G., Kubin M.Z. Functional analysis of TRAIL receptors using monoclonal antibodies. J. Immunol. 1999;162:2597–2605. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

