Isolation and Functional Characterization of MsFTa, a FLOWERING LOCUS T Homolog from Alfalfa (Medicago sativa)
- PMID: 31013631
- PMCID: PMC6514984
- DOI: 10.3390/ijms20081968
Isolation and Functional Characterization of MsFTa, a FLOWERING LOCUS T Homolog from Alfalfa (Medicago sativa)
Abstract
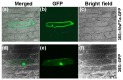
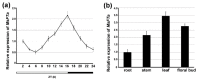
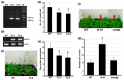
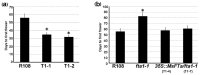
The production of hay and seeds of alfalfa, an important legume forage for the diary industry worldwide, is highly related to flowering time, which has been widely reported to be integrated by FLOWERING LOCUS T (FT). However, the function of FT(s) in alfalfa is largely unknown. Here, we identified MsFTa, an FT ortholog in alfalfa, and characterized its role in flowering regulation. MsFTa shares the conserved exon/intron structure of FTs, and MsFTa is 98% identical to MtFTa1 in Medicago trucatula. MsFTa was diurnally regulated with a peak before the dark period, and was preferentially expressed in leaves and floral buds. Transient expression of MsFTa-GFP fusion protein demonstrated its localization in the nucleus and cytoplasm. When ectopically expressed, MsFTa rescued the late-flowering phenotype of ft mutants from Arabidopsis and M. trucatula. MsFTa over-expression plants of both Arabidopsis and M. truncatula flowered significantly earlier than the non-transgenic controls under long day conditions, indicating that exogenous MsFTa strongly accelerated flowering. Hence, MsFTa functions positively in flowering promotion, suggesting that MsFTa may encode a florigen that acts as a key regulator in the flowering pathway. This study provides an effective candidate gene for optimizing alfalfa flowering time by genetically manipulating the expression of MsFTa.
Keywords: MsFTa; alfalfa; flowering time; transgenic plants.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time.Plant Physiol. 2011 Aug;156(4):2207-24. doi: 10.1104/pp.111.180182. Epub 2011 Jun 17. Plant Physiol. 2011. PMID: 21685176 Free PMC article.
-
Comparative analysis of the pteridophyte Adiantum MFT ortholog reveals the specificity of combined FT/MFT C and N terminal interaction with FD for the regulation of the downstream gene AP1.Plant Mol Biol. 2016 Jul;91(4-5):563-79. doi: 10.1007/s11103-016-0489-0. Epub 2016 May 23. Plant Mol Biol. 2016. PMID: 27216814
-
Expression of the alfalfa FRIGIDA-like gene, MsFRI-L delays flowering time in transgenic Arabidopsis thaliana.Mol Biol Rep. 2013 Mar;40(3):2083-90. doi: 10.1007/s11033-012-2266-8. Epub 2012 Dec 5. Mol Biol Rep. 2013. PMID: 23212611
-
The Use of Gene Modification and Advanced Molecular Structure Analyses towards Improving Alfalfa Forage.Int J Mol Sci. 2017 Jan 29;18(2):298. doi: 10.3390/ijms18020298. Int J Mol Sci. 2017. PMID: 28146083 Free PMC article. Review.
-
Biotechnological advancements in alfalfa improvement.J Appl Genet. 2011 May;52(2):111-24. doi: 10.1007/s13353-011-0028-2. Epub 2011 Jan 29. J Appl Genet. 2011. PMID: 21279557 Review.
Cited by
-
Multiplex CRISPR/Cas9-mediated mutagenesis of alfalfa FLOWERING LOCUS Ta1 (MsFTa1) leads to delayed flowering time with improved forage biomass yield and quality.Plant Biotechnol J. 2023 Jul;21(7):1383-1392. doi: 10.1111/pbi.14042. Epub 2023 Mar 25. Plant Biotechnol J. 2023. PMID: 36964962 Free PMC article.
-
Mechanisms of Vernalization-Induced Flowering in Legumes.Int J Mol Sci. 2022 Aug 31;23(17):9889. doi: 10.3390/ijms23179889. Int J Mol Sci. 2022. PMID: 36077286 Free PMC article. Review.
-
A Genome-Wide Association Study Coupled With a Transcriptomic Analysis Reveals the Genetic Loci and Candidate Genes Governing the Flowering Time in Alfalfa (Medicago sativa L.).Front Plant Sci. 2022 Jul 11;13:913947. doi: 10.3389/fpls.2022.913947. eCollection 2022. Front Plant Sci. 2022. PMID: 35898229 Free PMC article.
-
Identification and Functional Characterization of FLOWERING LOCUS T in Platycodon grandiflorus.Plants (Basel). 2022 Jan 26;11(3):325. doi: 10.3390/plants11030325. Plants (Basel). 2022. PMID: 35161306 Free PMC article.
-
Functional Characterization of the MsFKF1 Gene Reveals Its Dual Role in Regulating the Flowering Time and Plant Height in Medicago sativa L.Plants (Basel). 2024 Feb 27;13(5):655. doi: 10.3390/plants13050655. Plants (Basel). 2024. PMID: 38475501 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

