Tracking Biodistribution of Myeloid-Derived Cells in Murine Models of Breast Cancer
- PMID: 31013756
- PMCID: PMC6523772
- DOI: 10.3390/genes10040297
Tracking Biodistribution of Myeloid-Derived Cells in Murine Models of Breast Cancer
Abstract
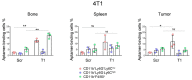
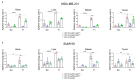
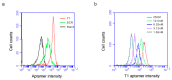
A growing tumor is constantly secreting inflammatory chemokines and cytokines that induce release of immature myeloid cells, including myeloid-derived suppressor cells (MDSCs) and macrophages, from the bone marrow. These cells not only promote tumor growth, but also prepare distant organs for tumor metastasis. On the other hand, the myeloid-derived cells also have phagocytic potential, and can serve as vehicles for drug delivery. We have previously identified thioaptamers that bind a subset of MDSCs with high affinity and specificity. In the current study, we applied one of the thioaptamers as a probe to track myeloid cell distribution in the bone, liver, spleen and tumor in multiple murine models of breast cancer including the 4T1 syngeneic model and MDA-MB-231 and SUM159 xenograft models. Information generated from this study will facilitate further understanding of tumor growth and metastasis, and predict biodistribution patterns of cell-mediated drug delivery.
Keywords: biodistribution; breast cancer; myeloid-derived suppressor cell; thioaptamer.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
CXCL17-derived CD11b+Gr-1+ myeloid-derived suppressor cells contribute to lung metastasis of breast cancer through platelet-derived growth factor-BB.Breast Cancer Res. 2019 Feb 12;21(1):23. doi: 10.1186/s13058-019-1114-3. Breast Cancer Res. 2019. PMID: 30755260 Free PMC article.
-
CCL9/CCR1 induces myeloid‑derived suppressor cell recruitment to the spleen in a murine H22 orthotopic hepatoma model.Oncol Rep. 2019 Jan;41(1):608-618. doi: 10.3892/or.2018.6809. Epub 2018 Oct 18. Oncol Rep. 2019. PMID: 30365155
-
Myeloid-derived suppressor cells accumulate among myeloid cells contributing to tumor growth in matrix metalloproteinase 12 knockout mice.Cell Immunol. 2018 May;327:1-12. doi: 10.1016/j.cellimm.2017.12.006. Epub 2017 Dec 13. Cell Immunol. 2018. PMID: 29555056
-
Suppressive role of myeloid-derived suppressor cells (MDSCs) in the microenvironment of breast cancer and targeted immunotherapies.Oncotarget. 2016 Sep 27;7(39):64505-64511. doi: 10.18632/oncotarget.11352. Oncotarget. 2016. PMID: 27542274 Free PMC article. Review.
-
Myeloid-derived suppressor cells: The green light for myeloma immune escape.Blood Rev. 2016 Sep;30(5):341-8. doi: 10.1016/j.blre.2016.04.002. Epub 2016 Apr 12. Blood Rev. 2016. PMID: 27132116 Free PMC article. Review.
References
-
- Bronte V., Brandau S., Chen S.H., Colombo M.P., Frey A.B., Greten T.F., Mandruzzato S., Murray P.J., Ochoa A., Ostrand-Rosenberg S., et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016;7:12150. doi: 10.1038/ncomms12150. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

