Transcription elements AREB6 and miR-34a affect apoptosis of PAMs by regulating the expression of SS2-related gene PPP1R11
- PMID: 31014175
- PMCID: PMC6527289
- DOI: 10.1080/15384101.2019.1610241
Transcription elements AREB6 and miR-34a affect apoptosis of PAMs by regulating the expression of SS2-related gene PPP1R11
Abstract
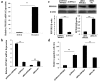
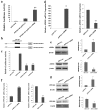
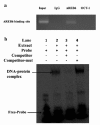
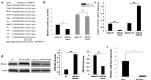
In our previous work, gene PPP1R11 (protein phosphatase 1 regulatory subunit 11) was significantly expressed in pigs after Streptococcus suis 2 (SS2) challenged. This study firstly confirmed that SS2 induced significant expression of PPP1R11 gene in porcine alveolar macrophage (PAM) cells, and apoptosis of PAM cells were observed. After that, the core promoter of porcine PPP1R11 was identified and its transcription factor AREB6 which significantly regulated PPP1R11. We also characterized that the PPP1R11 gene is a target of miR-34a. Further, we found that PPP1R11 helped to inhibit apoptosis of PAM cells under SS2 infecting, through transcription factor AREB6 was negatively correlated with apoptosis whereas miR-34a was positively correlated. Those findings provide a functional connection among the transcription factor AREB6, miR-34a, PPP1R11 gene and apoptosis of PAM cells in the pathogenesis of the SS2 infection.
Keywords: AREB6; Porcine alveolar macrophage; apoptosis; miR-34a.
Figures

Similar articles
-
MicroRNA-34b-5p targets PPP1R11 to inhibit proliferation and promote apoptosis in cattleyak Sertoli cells by regulating specific signaling pathways.Theriogenology. 2022 Dec;194:46-57. doi: 10.1016/j.theriogenology.2022.09.026. Epub 2022 Sep 29. Theriogenology. 2022. PMID: 36209544
-
Antagonistic Effects of p53 and HIF1A on microRNA-34a Regulation of PPP1R11 and STAT3 and Hypoxia-induced Epithelial to Mesenchymal Transition in Colorectal Cancer Cells.Gastroenterology. 2017 Aug;153(2):505-520. doi: 10.1053/j.gastro.2017.04.017. Epub 2017 Apr 20. Gastroenterology. 2017. PMID: 28435028
-
Investigation of Pathogenesis of H1N1 Influenza Virus and Swine Streptococcus suis Serotype 2 Co-Infection in Pigs by Microarray Analysis.PLoS One. 2015 Apr 23;10(4):e0124086. doi: 10.1371/journal.pone.0124086. eCollection 2015. PLoS One. 2015. PMID: 25906258 Free PMC article.
-
Viral Ubiquitin Ligase Stimulates Selective Host MicroRNA Expression by Targeting ZEB Transcriptional Repressors.Viruses. 2017 Aug 7;9(8):210. doi: 10.3390/v9080210. Viruses. 2017. PMID: 28783105 Free PMC article.
-
UBE2C, Directly Targeted by miR-548e-5p, Increases the Cellular Growth and Invasive Abilities of Cancer Cells Interacting with the EMT Marker Protein Zinc Finger E-box Binding Homeobox 1/2 in NSCLC.Theranostics. 2019 Mar 17;9(7):2036-2055. doi: 10.7150/thno.32738. eCollection 2019. Theranostics. 2019. Retraction in: Theranostics. 2020 Jul 25;10(21):9619. doi: 10.7150/thno.50254. PMID: 31037155 Free PMC article. Retracted.
References
-
- Gaur U, Xiong YY, Luo QP, et al. Breed-specific transcriptome response of spleen from six to eight week old piglet after infection with Streptococcus suis type 2. Mol Biol Rep. 2014;41:7865–7873. - PubMed
-
- Moorhead GB, Trinkle-Mulcahy L, Ulke-Lemee A.. Emerging roles of nuclear protein phosphatases. Nat Rev Mol Cell Biol. 2007;8:234–244. - PubMed
-
- Ceulemans H, Bollen M.. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84:1–39. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
