Identification of genomic regions associated with multi-silique trait in Brassica napus
- PMID: 31014236
- PMCID: PMC6480887
- DOI: 10.1186/s12864-019-5675-4
Identification of genomic regions associated with multi-silique trait in Brassica napus
Abstract
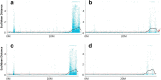
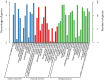
Background: Although rapeseed (Brassica napus L.) mutant forming multiple siliques was morphologically described and considered to increase the silique number per plant, an important agronomic trait in this crop, the molecular mechanism underlying this beneficial trait remains unclear. Here, we combined bulked-segregant analysis (BSA) and whole genome re-sequencing (WGR) to map the genomic regions responsible for the multi-silique trait using two pools of DNA from the near-isogenic lines (NILs) zws-ms (multi-silique) and zws-217 (single-silique). We used the Euclidean Distance (ED) to identify genomic regions associated with this trait based on both SNPs and InDels. We also conducted transcriptome sequencing to identify differentially expressed genes (DEGs) between zws-ms and zws-217.
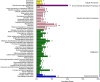
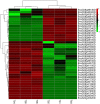
Results: Genetic analysis using the ED algorithm identified three SNP- and two InDel-associated regions for the multi-silique trait. Two highly overlapped parts of the SNP- and InDel-associated regions were identified as important intersecting regions, which are located on chromosomes A09 and C08, respectively, including 2044 genes in 10.20-MB length totally. Transcriptome sequencing revealed 129 DEGs between zws-ms and zws-217 in buds, including 39 DEGs located in the two abovementioned associated regions. We identified candidate genes involved in multi-silique formation in rapeseed based on the results of functional annotation.
Conclusions: This study identified the genomic regions and candidate genes related to the multi-silique trait in rapeseed.
Keywords: Association analysis; Brassica napus L.; Multi-silique; Near-isogenic line; Transcriptome sequencing; Whole genome re-sequencing.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Zhou HC, Jin L, Li J, Wang XJ. Altered callose deposition during embryo sac formation of multi-pistil mutant (mp1) in Medicago sativa. Genet Mol Res. 2016;15(2). 10.4238/gmr.15027968. - PubMed
-
- Li S, Yang L, Deng Q, Wang S, Wu F, Li P. Phenotypic characterization of a female sterile mutant in Rice. J Integr Plant Biol. 2006;48(3):307–314. doi: 10.1111/j.1744-7909.2006.00228.x. - DOI
MeSH terms
Grants and funding
- JFYS2016ZY03002156/The National Key Research and Development Plan
- 2018YFD0100500/The National Key Research and Development Plan
- CARS-12/Modern Agro-industry Technology Research System of China
- 09203020/Scientific Observing and Experimental Station of Oil Crops in the Upper Yangtze River, Ministry of Agriculture, P. R. China
- 2016ZYPZ-013/Financial Innovation Ability Promotion Project of Sichuan Province
LinkOut - more resources
Full Text Sources
Miscellaneous

