Estimates of introgression as a function of pairwise distances
- PMID: 31014244
- PMCID: PMC6480520
- DOI: 10.1186/s12859-019-2747-z
Estimates of introgression as a function of pairwise distances
Abstract
Background: Research over the last 10 years highlights the increasing importance of hybridization between species as a major force structuring the evolution of genomes and potentially providing raw material for adaptation by natural and/or sexual selection. Fueled by research in a few model systems where phenotypic hybrids are easily identified, research into hybridization and introgression (the flow of genes between species) has exploded with the advent of whole-genome sequencing and emerging methods to detect the signature of hybridization at the whole-genome or chromosome level. Amongst these are a general class of methods that utilize patterns of single-nucleotide polymorphisms (SNPs) across a tree as markers of hybridization. These methods have been applied to a variety of genomic systems ranging from butterflies to Neanderthals to detect introgression, however, when employed at a fine genomic scale these methods do not perform well to quantify introgression in small sample windows.
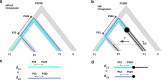
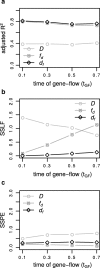
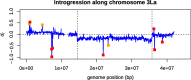
Results: We introduce a novel method to detect introgression by combining two widely used statistics: pairwise nucleotide diversity dxy and Patterson's D. The resulting statistic, the distance fraction (df), accounts for genetic distance across possible topologies and is designed to simultaneously detect and quantify introgression. We also relate our new method to the recently published fd and incorporate these statistics into the powerful genomics R-package PopGenome, freely available on GitHub (pievos101/PopGenome) and the Comprehensive R Archive Network (CRAN). The supplemental material contains a wide range of simulation studies and a detailed manual how to perform the statistics within the PopGenome framework.
Conclusion: We present a new distance based statistic df that avoids the pitfalls of Patterson's D when applied to small genomic regions and accurately quantifies the fraction of introgression (f) for a wide range of simulation scenarios.
Keywords: Genomics; Hybridisation; Introgression; SNPs.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Gilbert LE. Adaptive novelty through introgression in Heliconius wing patterns: evidence for shared genetic “tool box” from synthetic hybrid zones and a theory of diversification. In: Boggs CL, Watt W, Ehrlich P, editors. Ecology and Evolution Taking Flight: Butterflies as Model Systems. Chicago: University of Chicago Press; 2003.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

