Genomic sequence analysis reveals diversity of Australian Xanthomonas species associated with bacterial leaf spot of tomato, capsicum and chilli
- PMID: 31014247
- PMCID: PMC6480910
- DOI: 10.1186/s12864-019-5600-x
Genomic sequence analysis reveals diversity of Australian Xanthomonas species associated with bacterial leaf spot of tomato, capsicum and chilli
Abstract
Background: The genetic diversity in Australian populations of Xanthomonas species associated with bacterial leaf spot in tomato, capsicum and chilli were compared to worldwide bacterial populations. The aim of this study was to confirm the identities of these Australian Xanthomonas species and classify them in comparison to overseas isolates. Analysis of whole genome sequence allows for the investigation of bacterial population structure, pathogenicity and gene exchange, resulting in better management strategies and biosecurity.
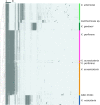
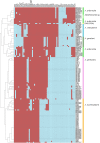
Results: Phylogenetic analysis of the core genome alignments and SNP data grouped strains in distinct clades. Patterns observed in average nucleotide identity, pan genome structure, effector and carbohydrate active enzyme profiles reflected the whole genome phylogeny and highlight taxonomic issues in X. perforans and X. euvesicatoria. Circular sequences with similarity to previously characterised plasmids were identified, and plasmids of similar sizes were isolated. Potential false positive and false negative plasmid assemblies were discussed. Effector patterns that may influence virulence on host plant species were analysed in pathogenic and non-pathogenic xanthomonads.
Conclusions: The phylogeny presented here confirmed X. vesicatoria, X. arboricola, X. euvesicatoria and X. perforans and a clade of an uncharacterised Xanthomonas species shown to be genetically distinct from all other strains of this study. The taxonomic status of X. perforans and X. euvesicatoria as one species is discussed in relation to whole genome phylogeny and phenotypic traits. The patterns evident in enzyme and plasmid profiles indicate worldwide exchange of genetic material with the potential to introduce new virulence elements into local bacterial populations.
Keywords: CAZymes; Cell wall degrading enzymes; Pan genome; Secretion system.
Conflict of interest statement
Ethics approval and consent to participate
NA
Consent for publication
NA
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- Potnis N, Timilsina S, Strayer A, Shantharaj D, Barak JD, Paret ML, Vallad GE, Jones JB. Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol Plant Pathol. 2015;16(9):907–920. doi: 10.1111/mpp.12244. - DOI - PMC - PubMed
-
- Garita-Cambronero J, Palacio-Bielsa A, Lopez MM, Cubero J. Pan-genomic analysis permits differentiation of virulent and non-virulent strains of Xanthomonas arboricola that cohabit Prunus spp and elucidate bacterial virulence factors. Front Microbiol. 2017;8:573. doi: 10.3389/fmicb.2017.00573. - DOI - PMC - PubMed
-
- Roach R, Mann R, Gambley CG, Shivas R, Rodoni B. Identification of Xanthomonas species associated with bacterial leaf spot of tomato, capsicum and chilli crops in eastern Australia. Eur J Plant Pathol. 2017;150(3):595–608. doi: 10.1007/s10658-017-1303-9. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

