Hes1 attenuates type I IFN responses via VEGF-C and WDFY1
- PMID: 31015298
- PMCID: PMC6547865
- DOI: 10.1084/jem.20180861
Hes1 attenuates type I IFN responses via VEGF-C and WDFY1
Abstract
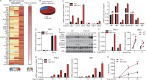
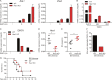
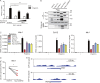
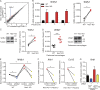
Induction of type I interferons (IFNs) is critical for eliciting competent immune responses, especially antiviral immunity. However, uncontrolled IFN production contributes to pathogenesis of autoimmune and inflammatory diseases. We found that transcription factor Hes1 suppressed production of type I IFNs and expression of IFN-stimulated genes. Functionally, Hes1-deficient mice displayed a heightened IFN signature in vivo, mounted enhanced resistance against encephalomyocarditis virus infection, and showed signs of exacerbated experimental lupus nephritis. Mechanistically, Hes1 did not suppress IFNs via direct transcriptional repression of IFN-encoding genes. Instead, Hes1 attenuated activation of TLR upstream signaling by inhibition of an adaptor molecule, WDFY1. Genome-wide assessment of Hes1 occupancy revealed that suppression of WDFY1 was secondary to direct binding and thus enhancement of expression of VEGF-C by Hes1, making Vegfc a rare example of an Hes1 positively regulated gene. In summary, these results identified Hes1 as a homeostatic negative regulator of type I IFNs for the maintenance of immune balance in the context of antiviral immunity and autoimmune diseases.
© 2019 Ning et al.
Figures


Similar articles
-
Scaffolding adaptor protein Gab1 is required for TLR3/4- and RIG-I-mediated production of proinflammatory cytokines and type I IFN in macrophages.J Immunol. 2010 Jun 1;184(11):6447-56. doi: 10.4049/jimmunol.0901750. Epub 2010 Apr 30. J Immunol. 2010. PMID: 20435932
-
Type I Interferon response in olfactory bulb, the site of tick-borne flavivirus accumulation, is primarily regulated by IPS-1.J Neuroinflammation. 2016 Jan 27;13:22. doi: 10.1186/s12974-016-0487-9. J Neuroinflammation. 2016. PMID: 26819220 Free PMC article.
-
The transcriptional repressor Hes1 attenuates inflammation by regulating transcription elongation.Nat Immunol. 2016 Aug;17(8):930-7. doi: 10.1038/ni.3486. Epub 2016 Jun 20. Nat Immunol. 2016. PMID: 27322654 Free PMC article.
-
Regulation of type I interferon signaling in immunity and inflammation: A comprehensive review.J Autoimmun. 2017 Sep;83:1-11. doi: 10.1016/j.jaut.2017.03.008. Epub 2017 Mar 19. J Autoimmun. 2017. PMID: 28330758 Review.
-
TICAM-1 and TICAM-2: toll-like receptor adapters that participate in induction of type 1 interferons.Int J Biochem Cell Biol. 2005 Mar;37(3):524-9. doi: 10.1016/j.biocel.2004.07.018. Int J Biochem Cell Biol. 2005. PMID: 15618008 Review.
Cited by
-
Secondary metabolites of Bacillus subtilis L2 show antiviral activity against pseudorabies virus.Front Microbiol. 2023 Oct 30;14:1277782. doi: 10.3389/fmicb.2023.1277782. eCollection 2023. Front Microbiol. 2023. PMID: 37965547 Free PMC article.
-
Pseudorabies virus tegument protein UL13 recruits RNF5 to inhibit STING-mediated antiviral immunity.PLoS Pathog. 2022 May 18;18(5):e1010544. doi: 10.1371/journal.ppat.1010544. eCollection 2022 May. PLoS Pathog. 2022. PMID: 35584187 Free PMC article.
-
The HDAC inhibitor domatinostat induces type I interferon α in Merkel cell carcinoma by HES1 repression.J Cancer Res Clin Oncol. 2023 Sep;149(11):8267-8277. doi: 10.1007/s00432-023-04733-y. Epub 2023 Apr 18. J Cancer Res Clin Oncol. 2023. PMID: 37071208 Free PMC article.
-
Evaluation of large language models for discovery of gene set function.ArXiv [Preprint]. 2024 Apr 1:arXiv:2309.04019v2. ArXiv. 2024. Update in: Nat Methods. 2025 Jan;22(1):82-91. doi: 10.1038/s41592-024-02525-x. PMID: 37731657 Free PMC article. Updated. Preprint.
-
Pseudorabies Virus Tegument Protein UL13 Suppresses RLR-Mediated Antiviral Innate Immunity through Regulating Receptor Transcription.Viruses. 2022 Jul 2;14(7):1465. doi: 10.3390/v14071465. Viruses. 2022. PMID: 35891444 Free PMC article.
References
-
- Alvarez Y., Municio C., Hugo E., Zhu J., Alonso S., Hu X., Fernández N., and Sánchez Crespo M.. 2011. Notch- and transducin-like enhancer of split (TLE)-dependent histone deacetylation explain interleukin 12 (IL-12) p70 inhibition by zymosan. J. Biol. Chem. 286:16583–16595. 10.1074/jbc.M111.222158 - DOI - PMC - PubMed
-
- Arisi I., D’Onofrio M., Brandi R., Felsani A., Capsoni S., Drovandi G., Felici G., Weitschek E., Bertolazzi P., and Cattaneo A.. 2011. Gene expression biomarkers in the brain of a mouse model for Alzheimer’s disease: mining of microarray data by logic classification and feature selection. J. Alzheimers Dis. 24:721–738. 10.3233/JAD-2011-101881 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

