PARP Inhibitor Efficacy Depends on CD8+ T-cell Recruitment via Intratumoral STING Pathway Activation in BRCA-Deficient Models of Triple-Negative Breast Cancer
- PMID: 31015319
- PMCID: PMC6548644
- DOI: 10.1158/2159-8290.CD-18-1218
PARP Inhibitor Efficacy Depends on CD8+ T-cell Recruitment via Intratumoral STING Pathway Activation in BRCA-Deficient Models of Triple-Negative Breast Cancer
Abstract
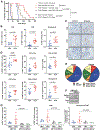
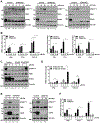
Combinatorial clinical trials of PARP inhibitors with immunotherapies are ongoing, yet the immunomodulatory effects of PARP inhibition have been incompletely studied. Here, we sought to dissect the mechanisms underlying PARP inhibitor-induced changes in the tumor microenvironment of BRCA1-deficient triple-negative breast cancer (TNBC). We demonstrate that the PARP inhibitor olaparib induces CD8+ T-cell infiltration and activation in vivo, and that CD8+ T-cell depletion severely compromises antitumor efficacy. Olaparib-induced T-cell recruitment is mediated through activation of the cGAS/STING pathway in tumor cells with paracrine activation of dendritic cells and is more pronounced in HR-deficient compared with HR-proficient TNBC cells and in vivo models. CRISPR-mediated knockout of STING in cancer cells prevents proinflammatory signaling and is sufficient to abolish olaparib-induced T-cell infiltration in vivo. These findings elucidate an additional mechanism of action of PARP inhibitors and provide a rationale for combining PARP inhibition with immunotherapies for the treatment of TNBC. SIGNIFICANCE: This work demonstrates cross-talk between PARP inhibition and the tumor microenvironment related to STING/TBK1/IRF3 pathway activation in cancer cells that governs CD8+ T-cell recruitment and antitumor efficacy. The data provide insight into the mechanism of action of PARP inhibitors in BRCA-associated breast cancer.This article is highlighted in the In This Issue feature, p. 681.
©2019 American Association for Cancer Research.
Figures




References
-
- Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol 2010;7:683–92. - PubMed
-
- Robson M, Im S-A, Senkus E, Xu B, mchek S, Masuda N, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. New Engl J Med 2017;377:523–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

