Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli
- PMID: 31015395
- PMCID: PMC6478713
- DOI: 10.1038/s41467-019-09507-0
Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli
Abstract
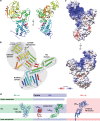
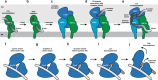
The bacterial cell wall plays a crucial role in viability and is an important drug target. In Escherichia coli, the peptidoglycan crosslinking reaction to form the cell wall is primarily carried out by penicillin-binding proteins that catalyse D,D-transpeptidase activity. However, an alternate crosslinking mechanism involving the L,D-transpeptidase YcbB can lead to bypass of D,D-transpeptidation and beta-lactam resistance. Here, we show that the crystallographic structure of YcbB consists of a conserved L,D-transpeptidase catalytic domain decorated with a subdomain on the dynamic substrate capping loop, peptidoglycan-binding and large scaffolding domains. Meropenem acylation of YcbB gives insight into the mode of inhibition by carbapenems, the singular antibiotic class with significant activity against L,D-transpeptidases. We also report the structure of PBP5-meropenem to compare interactions mediating inhibition. Additionally, we probe the interaction network of this pathway and assay beta-lactam resistance in vivo. Our results provide structural insights into the mechanism of action and the inhibition of L,D-transpeptidation, and into YcbB-mediated antibiotic resistance.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

