Corrupted coordination of epigenetic modifications leads to diverging chromatin states and transcriptional heterogeneity in CLL
- PMID: 31015400
- PMCID: PMC6478836
- DOI: 10.1038/s41467-019-09645-5
Corrupted coordination of epigenetic modifications leads to diverging chromatin states and transcriptional heterogeneity in CLL
Abstract
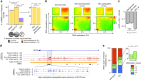
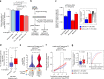
Cancer evolution is fueled by epigenetic as well as genetic diversity. In chronic lymphocytic leukemia (CLL), intra-tumoral DNA methylation (DNAme) heterogeneity empowers evolution. Here, to comprehensively study the epigenetic dimension of cancer evolution, we integrate DNAme analysis with histone modification mapping and single cell analyses of RNA expression and DNAme in 22 primary CLL and 13 healthy donor B lymphocyte samples. Our data reveal corrupted coherence across different layers of the CLL epigenome. This manifests in decreased mutual information across epigenetic modifications and gene expression attributed to cell-to-cell heterogeneity. Disrupted epigenetic-transcriptional coordination in CLL is also reflected in the dysregulation of the transcriptional output as a function of the combinatorial chromatin states, including incomplete Polycomb-mediated gene silencing. Notably, we observe unexpected co-mapping of typically mutually exclusive activating and repressing histone modifications, suggestive of intra-tumoral epigenetic diversity. Thus, CLL epigenetic diversification leads to decreased coordination across layers of epigenetic information, likely reflecting an admixture of cells with diverging cellular identities.
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Epigenetic cancer evolution, one cell at a time.Nat Rev Genet. 2019 Aug;20(8):434-435. doi: 10.1038/s41576-019-0143-1. Nat Rev Genet. 2019. PMID: 31160791 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

