The H2A.Z histone variant integrates Wnt signaling in intestinal epithelial homeostasis
- PMID: 31015444
- PMCID: PMC6478875
- DOI: 10.1038/s41467-019-09899-z
The H2A.Z histone variant integrates Wnt signaling in intestinal epithelial homeostasis
Abstract
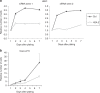
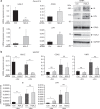
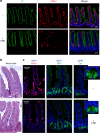
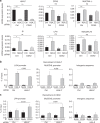
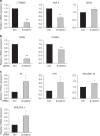
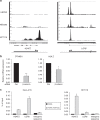
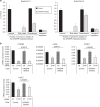
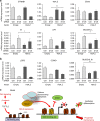
The Tip60/p400 chromatin-modifying complex, which is involved in the incorporation and post-translational modification of the H2A.Z histone variant, regulates cell proliferation and important signaling pathways, such as Wnt. Here, we study the involvement of H2A.Z in intestinal epithelial homeostasis, which is dependent on the finely-tuned equilibrium between stem cells renewal and differentiation, under the control of such pathway. We use cell models and inducible knock-out mice to study the impact of H2A.Z depletion on intestinal homeostasis. We show that H2A.Z is essential for the proliferation of human cancer and normal intestinal crypt cells and negatively controls the expression of a subset of differentiation markers, in cultured cells and mice. H2A.Z impairs the recruitment of the intestine-specific transcription factor CDX2 to chromatin, is itself a target of the Wnt pathway and thus, acts as an integrator for Wnt signaling in the control of intestinal epithelial cell fate and homeostasis.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

