Multiple myeloma immunoglobulin lambda translocations portend poor prognosis
- PMID: 31015454
- PMCID: PMC6478743
- DOI: 10.1038/s41467-019-09555-6
Multiple myeloma immunoglobulin lambda translocations portend poor prognosis
Abstract
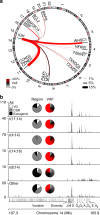
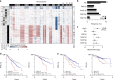
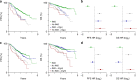
Multiple myeloma is a malignancy of antibody-secreting plasma cells. Most patients benefit from current therapies, however, 20% of patients relapse or die within two years and are deemed high risk. Here we analyze structural variants from 795 newly-diagnosed patients as part of the CoMMpass study. We report translocations involving the immunoglobulin lambda (IgL) locus are present in 10% of patients, and indicative of poor prognosis. This is particularly true for IgL-MYC translocations, which coincide with focal amplifications of enhancers at both loci. Importantly, 78% of IgL-MYC translocations co-occur with hyperdiploid disease, a marker of standard risk, suggesting that IgL-MYC-translocated myeloma is being misclassified. Patients with IgL-translocations fail to benefit from IMiDs, which target IKZF1, a transcription factor that binds the IgL enhancer at some of the highest levels in the myeloma epigenome. These data implicate IgL translocation as a driver of poor prognosis which may be due to IMiD resistance.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

