K27-linked ubiquitination of BRAF by ITCH engages cytokine response to maintain MEK-ERK signaling
- PMID: 31015455
- PMCID: PMC6478693
- DOI: 10.1038/s41467-019-09844-0
K27-linked ubiquitination of BRAF by ITCH engages cytokine response to maintain MEK-ERK signaling
Abstract
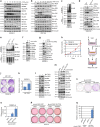
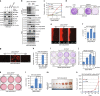
BRAF plays an indispensable role in activating the MEK/ERK pathway to drive tumorigenesis. Receptor tyrosine kinase and RAS-mediated BRAF activation have been extensively characterized, however, it remains undefined how BRAF function is fine-tuned by stimuli other than growth factors. Here, we report that in response to proinflammatory cytokines, BRAF is subjected to lysine 27-linked poly-ubiquitination in melanoma cells by the ITCH ubiquitin E3 ligase. Lysine 27-linked ubiquitination of BRAF recruits PP2A to antagonize the S365 phosphorylation and disrupts the inhibitory interaction with 14-3-3, leading to sustained BRAF activation and subsequent elevation of the MEK/ERK signaling. Physiologically, proinflammatory cytokines activate ITCH to maintain BRAF activity and to promote proliferation and invasion of melanoma cells, whereas the ubiquitination-deficient BRAF mutant displays compromised kinase activity and reduced tumorigenicity. Collectively, our study reveals a pivotal role for ITCH-mediated BRAF ubiquitination in coordinating the signals between cytokines and the MAPK pathway activation in melanoma cells.
Conflict of interest statement
The authors declare no competing interests.
Figures

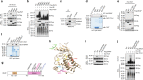



Similar articles
-
The APC/C E3 Ligase Complex Activator FZR1 Restricts BRAF Oncogenic Function.Cancer Discov. 2017 Apr;7(4):424-441. doi: 10.1158/2159-8290.CD-16-0647. Epub 2017 Feb 7. Cancer Discov. 2017. PMID: 28174173 Free PMC article.
-
The BRAF(V600E) inhibitor, PLX4032, increases type I collagen synthesis in melanoma cells.Matrix Biol. 2015 Oct;48:66-77. doi: 10.1016/j.matbio.2015.05.007. Epub 2015 May 16. Matrix Biol. 2015. PMID: 25989506 Free PMC article.
-
Antitumor efficacy of the novel RAF inhibitor GDC-0879 is predicted by BRAFV600E mutational status and sustained extracellular signal-regulated kinase/mitogen-activated protein kinase pathway suppression.Cancer Res. 2009 Apr 1;69(7):3042-51. doi: 10.1158/0008-5472.CAN-08-3563. Epub 2009 Mar 10. Cancer Res. 2009. PMID: 19276360
-
MEK and RAF inhibitors for BRAF-mutated cancers.Expert Rev Mol Med. 2012 Oct 12;14:e17. doi: 10.1017/erm.2012.11. Expert Rev Mol Med. 2012. PMID: 23058743 Review.
-
A pivotal role for ERK in the oncogenic behaviour of malignant melanoma?Int J Cancer. 2003 May 1;104(5):527-32. doi: 10.1002/ijc.10978. Int J Cancer. 2003. PMID: 12594806 Review.
Cited by
-
The mTOR pathway controls phosphorylation of BRAF at T401.Cell Commun Signal. 2024 Sep 2;22(1):428. doi: 10.1186/s12964-024-01808-2. Cell Commun Signal. 2024. PMID: 39223665 Free PMC article.
-
ITCH as a potential therapeutic target in human cancers.Semin Cancer Biol. 2020 Dec;67(Pt 2):117-130. doi: 10.1016/j.semcancer.2020.03.003. Epub 2020 Mar 10. Semin Cancer Biol. 2020. PMID: 32165318 Free PMC article. Review.
-
The combined signatures of the tumour microenvironment and nucleotide metabolism-related genes provide a prognostic and therapeutic biomarker for gastric cancer.Sci Rep. 2023 Apr 24;13(1):6622. doi: 10.1038/s41598-023-33213-z. Sci Rep. 2023. PMID: 37095256 Free PMC article.
-
The E3 ubiquitin ligase Itch regulates death receptor and cholesterol trafficking to affect TRAIL-mediated apoptosis.Cell Death Dis. 2024 Jan 12;15(1):40. doi: 10.1038/s41419-023-06417-4. Cell Death Dis. 2024. PMID: 38216558 Free PMC article.
-
TRIM21-mediated ubiquitination and phosphorylation of ERK1/2 promotes cell proliferation and drug resistance in pituitary adenomas.Neuro Oncol. 2025 Mar 7;27(3):727-742. doi: 10.1093/neuonc/noae241. Neuro Oncol. 2025. PMID: 39533840
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

