Phenotypic characteristics and transcriptome profile of Cryptococcus gattii biofilm
- PMID: 31015652
- PMCID: PMC6478838
- DOI: 10.1038/s41598-019-42896-2
Phenotypic characteristics and transcriptome profile of Cryptococcus gattii biofilm
Abstract
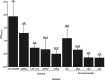
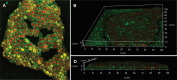
In this study, we characterized Cryptococcus gattii biofilm formation in vitro. There was an increase in the density of metabolically active sessile cells up to 72 h of biofilm formation on polystyrene and glass surfaces. Scanning electron microscopy and confocal laser scanning microscopy analysis revealed that in the early stage of biofilm formation, yeast cells adhered to the abiotic surface as a monolayer. After 12 h, extracellular fibrils were observed projecting from C. gattii cells, connecting the yeast cells to each other and to the abiotic surface; mature biofilm consisted of a dense network of cells deeply encased in an extracellular polymeric matrix. These features were also observed in biofilms formed on polyvinyl chloride and silicone catheter surfaces. We used RNA-Seq-based transcriptome analysis to identify changes in gene expression associated with C. gattii biofilm at 48 h compared to the free-floating planktonic cells. Differential expression analysis showed that 97 and 224 transcripts were up-regulated and down-regulated in biofilm, respectively. Among the biological processes, the highest enriched term showed that the transcripts were associated with cellular metabolic processes, macromolecule biosynthetic processes and translation.
Conflict of interest statement
The authors declare no competing interests.
Figures


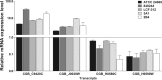
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

