Heterologous transporters from anaerobic fungi bolster fluoride tolerance in Saccharomyces cerevisiae
- PMID: 31016136
- PMCID: PMC6475669
- DOI: 10.1016/j.mec.2019.e00091
Heterologous transporters from anaerobic fungi bolster fluoride tolerance in Saccharomyces cerevisiae
Abstract
Membrane-embedded transporters are crucial for the stability and performance of microbial production strains. Apart from engineering known transporters derived from model systems, it is equally important to identify transporters from nonconventional organisms that confer advantageous traits for biotechnological applications. Here, we transferred genes encoding fluoride exporter (FEX) proteins from three strains of early-branching anaerobic fungi (Neocallimastigomycota) to Saccharomyces cerevisiae. The heterologous transporters are localized to the plasma membrane and complement a fluoride-sensitive yeast strain that is lacking endogenous fluoride transporters up to 10.24 mM fluoride. Furthermore, we show that fusing an amino-terminal leader sequence to FEX proteins in yeast elevates protein yields, yet inadvertently causes a loss of transporter function. Adaptive laboratory evolution of FEX proteins restores fluoride tolerance of these strains, in one case exceeding the solute tolerance observed in wild type S. cerevisiae; however, the underlying molecular mechanisms and cause for the increased tolerance in the evolved strains remain elusive. Our results suggest that microbial cultures can achieve solvent tolerance through different adaptive trajectories, and the study is a promising step towards the identification, production, and biotechnological application of membrane proteins from nonconventional fungi.
Keywords: Anaerobic gut fungi; Fluoride export proteins; Membrane proteins; Microbial engineering; Neocallimastigomycota.
Figures

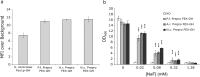


Similar articles
-
A SWEET surprise: Anaerobic fungal sugar transporters and chimeras enhance sugar uptake in yeast.Metab Eng. 2021 Jul;66:137-147. doi: 10.1016/j.ymben.2021.04.009. Epub 2021 Apr 19. Metab Eng. 2021. PMID: 33887459
-
The fluoride transporter FLUORIDE EXPORTER (FEX) is the major mechanism of tolerance to fluoride toxicity in plants1.Plant Physiol. 2021 Mar 21:kiab131. doi: 10.1093/plphys/kiab131. Online ahead of print. Plant Physiol. 2021. PMID: 33787927
-
The fluoride transporter FLUORIDE EXPORTER (FEX) is the major mechanism of tolerance to fluoride toxicity in plants.Plant Physiol. 2021 Mar 21;186(2):1143-58. doi: 10.1093/plphys/kiab131. Online ahead of print. Plant Physiol. 2021. PMID: 33744970 Free PMC article.
-
Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential.FEMS Microbiol Ecol. 2014 Oct;90(1):1-17. doi: 10.1111/1574-6941.12383. Epub 2014 Aug 11. FEMS Microbiol Ecol. 2014. PMID: 25046344 Review.
-
Engineering of Pentose Transport in Saccharomyces cerevisiae for Biotechnological Applications.Front Bioeng Biotechnol. 2020 Jan 29;7:464. doi: 10.3389/fbioe.2019.00464. eCollection 2019. Front Bioeng Biotechnol. 2020. PMID: 32064252 Free PMC article. Review.
Cited by
-
Co‑cultivation of the anaerobic fungus Caecomyces churrovis with Methanobacterium bryantii enhances transcription of carbohydrate binding modules, dockerins, and pyruvate formate lyases on specific substrates.Biotechnol Biofuels. 2021 Dec 10;14(1):234. doi: 10.1186/s13068-021-02083-w. Biotechnol Biofuels. 2021. PMID: 34893091 Free PMC article.
-
The Anaerobic Fungi: Challenges and Opportunities for Industrial Lignocellulosic Biofuel Production.Microorganisms. 2021 Mar 27;9(4):694. doi: 10.3390/microorganisms9040694. Microorganisms. 2021. PMID: 33801700 Free PMC article. Review.
-
Enzyme Discovery in Anaerobic Fungi (Neocallimastigomycetes) Enables Lignocellulosic Biorefinery Innovation.Microbiol Mol Biol Rev. 2022 Dec 21;86(4):e0004122. doi: 10.1128/mmbr.00041-22. Epub 2022 Jul 19. Microbiol Mol Biol Rev. 2022. PMID: 35852448 Free PMC article. Review.
-
Anaerobic Fungal Mevalonate Pathway Genomic Biases Lead to Heterologous Toxicity Underpredicted by Codon Adaptation Indices.Microorganisms. 2021 Sep 18;9(9):1986. doi: 10.3390/microorganisms9091986. Microorganisms. 2021. PMID: 34576881 Free PMC article.
-
A Genetic Engineering Toolbox for the Lignocellulolytic Anaerobic Gut Fungus Neocallimastix frontalis.ACS Synth Biol. 2023 Apr 21;12(4):1034-1045. doi: 10.1021/acssynbio.2c00502. Epub 2023 Mar 15. ACS Synth Biol. 2023. PMID: 36920337 Free PMC article.
References
-
- Arnold C.E., Parekh R.N., Yang W., Wittrup K.D. vol 104. 1998. (Leader Peptide Efficiency Correlates with Signal Recognition Particle Dependence in Saccharomyces cerevisiae). - PubMed
-
- Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E., Grosdidier A., Hernandez C., Ioannidis V., Kuznetsov D., Liechti R., Moretti S., Mostaguir K., Redaschi N., Rossier G., Xenarios I., Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

