Identification of a complex genomic rearrangement in TMPRSS3 by massively parallel sequencing in Chinese cases with prelingual hearing loss
- PMID: 31016883
- PMCID: PMC6565588
- DOI: 10.1002/mgg3.685
Identification of a complex genomic rearrangement in TMPRSS3 by massively parallel sequencing in Chinese cases with prelingual hearing loss
Abstract
Background: Genetic variants in TMPRSS3 have been causally linked to autosomal recessive nonsyndromic hearing loss (HL) at the DFNB8 and DFNB10 loci. These variants include both single nucleotide and copy number variations (CNVs). In this study, we aim to identify the genetic cause in three Chinese subjects with prelingual profound sensorineural HL.
Methods: We applied targeted genomic enrichment and massively parallel sequencing to screen 110 genes associated with nonsyndromic HL in the three affected subjects. CNVplex® analysis and polymerase chain reaction (PCR) were performed for CNV detection.
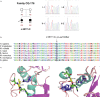
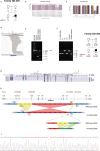
Results: We identified biallelic variations in TMPRSS3 including a novel complex genomic rearrangement and a novel missense mutation, c.551T>C. We have mapped the breakpoints of the genomic rearrangement and showed that it consisted of two deletions and an inversion encompassing exon 3 to exon 9 of TMPRSS3.
Conclusion: Our study expanded the mutational spectrum of TMPRSS3 to include complex genomic rearrangements. It showcased the importance of an integrative approach to investigate CNVs and their contribution to HL.
Keywords: TMPRSS3; copy number variation; hearing loss; massively parallel sequencing; rearrangement.
© 2019 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Conflict of interest statement
None declared.
Figures

Similar articles
-
Genotype-Phenotype Correlations in TMPRSS3 (DFNB10/DFNB8) with Emphasis on Natural History.Audiol Neurootol. 2023;28(6):407-419. doi: 10.1159/000528766. Epub 2023 Jun 16. Audiol Neurootol. 2023. PMID: 37331337 Free PMC article. Review.
-
Identification of TMPRSS3 as a Significant Contributor to Autosomal Recessive Hearing Loss in the Chinese Population.Neural Plast. 2017;2017:3192090. doi: 10.1155/2017/3192090. Epub 2017 Jun 13. Neural Plast. 2017. PMID: 28695016 Free PMC article.
-
A novel mutation of TMPRSS3 related to milder auditory phenotype in Korean postlingual deafness: a possible future implication for a personalized auditory rehabilitation.J Mol Med (Berl). 2014 Jun;92(6):651-63. doi: 10.1007/s00109-014-1128-3. Epub 2014 Feb 15. J Mol Med (Berl). 2014. PMID: 24526180
-
Novel Mutations and Mutation Combinations of TMPRSS3 Cause Various Phenotypes in One Chinese Family with Autosomal Recessive Hearing Impairment.Biomed Res Int. 2017;2017:4707315. doi: 10.1155/2017/4707315. Epub 2017 Jan 29. Biomed Res Int. 2017. PMID: 28246597 Free PMC article.
-
TMPRSS3, a type II transmembrane serine protease mutated in non-syndromic autosomal recessive deafness.Front Biosci. 2008 Jan 1;13:1557-67. doi: 10.2741/2780. Front Biosci. 2008. PMID: 17981648 Review.
Cited by
-
Genotype-Phenotype Correlations in TMPRSS3 (DFNB10/DFNB8) with Emphasis on Natural History.Audiol Neurootol. 2023;28(6):407-419. doi: 10.1159/000528766. Epub 2023 Jun 16. Audiol Neurootol. 2023. PMID: 37331337 Free PMC article. Review.
-
TMPRSS3 Gene Variants With Implications for Auditory Treatment and Counseling.Front Genet. 2021 Nov 19;12:780874. doi: 10.3389/fgene.2021.780874. eCollection 2021. Front Genet. 2021. PMID: 34868270 Free PMC article.
-
[Late-onset hereditary hearing loss caused by TMPRSS3 compound heterozygous mutations].Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2024 Aug;38(8):679-686. doi: 10.13201/j.issn.2096-7993.2024.08.002. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2024. PMID: 39118504 Free PMC article. Chinese.
References
-
- Azaiez, H. , Booth, K. T. , Ephraim, S. S. , Crone, B. , Black‐Ziegelbein, E. A. , Marini, R. J. , … Smith, R. J. H. (2018). Genomic landscape and mutational signatures of deafness‐associated genes. The American Journal of Human Genetics, 103(4), 484–497. 10.1016/j.ajhg.2018.08.006 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

