Analysis of a nanoparticle‑enriched fraction of plasma reveals miRNA candidates for Down syndrome pathogenesis
- PMID: 31017260
- PMCID: PMC6488180
- DOI: 10.3892/ijmm.2019.4158
Analysis of a nanoparticle‑enriched fraction of plasma reveals miRNA candidates for Down syndrome pathogenesis
Erratum in
-
[Corrigendum] Analysis of a nanoparticle‑enriched fraction of plasma reveals miRNA candidates for Down syndrome pathogenesis.Int J Mol Med. 2019 Aug;44(2):768. doi: 10.3892/ijmm.2019.4222. Epub 2019 May 30. Int J Mol Med. 2019. PMID: 31173160 Free PMC article.
Abstract
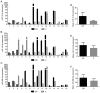
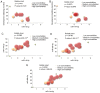
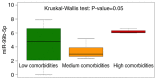
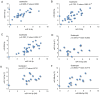
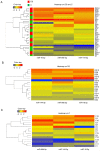
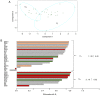
Down syndrome (DS) is caused by the presence of part or all of a third copy of chromosome 21. DS is associated with several phenotypes, including intellectual disability, congenital heart disease, childhood leukemia and immune defects. Specific microRNAs (miRNAs/miR) have been described to be associated with DS, although none of them so far have been unequivocally linked to the pathology. The present study focuses to the best of our knowledge for the first time on the miRNAs contained in nanosized RNA carriers circulating in the blood. Fractions enriched in nanosized RNA‑carriers were separated from the plasma of young participants with DS and their non‑trisomic siblings and miRNAs were extracted. A microarray‑based analysis on a small cohort of samples led to the identification of the three most abundant miRNAs, namely miR‑16‑5p, miR‑99b‑5p and miR‑144‑3p. These miRNAs were then profiled for 15 pairs of DS and non‑trisomic sibling couples by reverse transcription‑quantitative polymerase chain reaction (RT‑qPCR). Results identified a clear differential expression trend of these miRNAs in DS with respect to their non‑trisomic siblings and gene ontology analysis pointed to their potential role in a number of typical DS features, including 'nervous system development', 'neuronal cell body' and certain forms of 'leukemia'. Finally, these expression levels were associated with certain typical quantitative and qualitative clinical features of DS. These results contribute to the efforts in defining the DS‑associated pathogenic mechanisms and emphasize the importance of properly stratifying the miRNA fluid vehicles in order to probe biomolecules that are otherwise hidden and/or not accessible to (standard) analysis.
Figures

References
-
- Strippoli P, Pelleri MC, Caracausi M, Vitale L, Piovesan A, Locatelli C, Mimmi MC, Berardi AC, Ricotta D, Radeghieri A, et al. An integrated route to identifying new pathogenesis-based therapeutic approaches for trisomy 21 (Down Syndrome) following the thought of Jérôme Lejeune. Sci Postprint. 2013;1:e00010. doi: 10.14340/spp.2013.12R0005. - DOI
-
- Delabar JM, Allinquant B, Bianchi D, Blumenthal T, Dekker A, Edgin J, O'Bryan J, Dierssen M, Potier MC, Wiseman F, et al. Changing paradigms in down syndrome: The first international conference of the trisomy 21 research society. Mol Syndromol. 2016;7:251–261. doi: 10.1159/000449049. - DOI - PMC - PubMed
-
- Lejeune J, Gauthier M, Turpin R. Human chromosomes in tissue cultures. C R Hebd Seances Acad Sci. 1959;248:602–603. In French. - PubMed
-
- Chen YQ, Li T, Guo WY, Su FJ, Zhang YX. Identification of altered pathways in down syndrome-associated congenital heart defects using an individualized pathway aberrance score. Genet Mol Res. 2016;15 - PubMed
-
- Pelleri MC, Gennari E, Locatelli C, Piovesan A, Caracausi M, Antonaros F, Rocca A, Donati CM, Conti L, Strippoli P, et al. Genotype-phenotype correlation for congenital heart disease in down syndrome through analysis of partial trisomy 21 cases. Genomics. 2017;109:391–400. doi: 10.1016/j.ygeno.2017.06.004. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

