A context-based ABC model for literature-based discovery
- PMID: 31017923
- PMCID: PMC6481912
- DOI: 10.1371/journal.pone.0215313
A context-based ABC model for literature-based discovery
Abstract
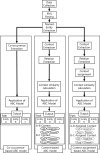
Background: In the literature-based discovery, considerable research has been done based on the ABC model developed by Swanson. ABC model hypothesizes that there is a meaningful relation between entity A extracted from document set 1 and entity C extracted from document set 2 through B entities that appear commonly in both document sets. The results of ABC model are relations among entity A, B, and C, which is referred as paths. A path allows for hypothesizing the relationship between entity A and entity C, or helps discover entity B as a new evidence for the relationship between entity A and entity C. The co-occurrence based approach of ABC model is a well-known approach to automatic hypothesis generation by creating various paths. However, the co-occurrence based ABC model has a limitation, in that biological context is not considered. It focuses only on matching of B entity which commonly appears in relation between two entities. Therefore, the paths extracted by the co-occurrence based ABC model tend to include a lot of irrelevant paths, meaning that expert verification is essential.
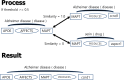
Methods: In order to overcome this limitation of the co-occurrence based ABC model, we propose a context-based approach to connecting one entity relation to another, modifying the ABC model using biological contexts. In this study, we defined four biological context elements: cell, drug, disease, and organism. Based on these biological context, we propose two extended ABC models: a context-based ABC model and a context-assignment-based ABC model. In order to measure the performance of the both proposed models, we examined the relevance of the B entities between the well-known relations "APOE-MAPT" as well as "FUS-TARDBP". Each relation means interaction between neurodegenerative disease associated with proteins. The interaction between APOE and MAPT is known to play a crucial role in Alzheimer's disease as APOE affects tau-mediated neurodegeneration. It has been shown that mutation in FUS and TARDBP are associated with amyotrophic lateral sclerosis(ALS), a motor neuron disease by leading to neuronal cell death. Using these two relations, we compared both of proposed models to co-occurrence based ABC model.
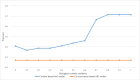
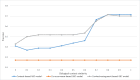
Results: The precision of B entities by co-occurrence based ABC model was 27.1% for "APOE-MAPT" and 22.1% for "FUS-TARDBP", respectively. In context-based ABC model, precision of extracted B entities was 71.4% for "APOE-MAPT", and 77.9% for "FUS-TARDBP". Context-assignment based ABC model achieved 89% and 97.5% precision for the two relations, respectively. Both proposed models achieved a higher precision than co-occurrence-based ABC model.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Swanson DR. Undiscovered public knowledge. The Library Quarterly. 1986;56(2):103–118.
-
- Leroy G, Chen H. Genescene: An ontology-enhanced integration of linguistic and co-occurrence based relations in biomedical texts. Journal of the American Society for Information Science and Technology. 2005;56(5):457–468.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

