PAD2-Mediated Citrullination Contributes to Efficient Oligodendrocyte Differentiation and Myelination
- PMID: 31018126
- PMCID: PMC6486480
- DOI: 10.1016/j.celrep.2019.03.108
PAD2-Mediated Citrullination Contributes to Efficient Oligodendrocyte Differentiation and Myelination
Abstract
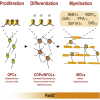
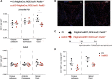
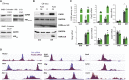
Citrullination, the deimination of peptidylarginine residues into peptidylcitrulline, has been implicated in the etiology of several diseases. In multiple sclerosis, citrullination is thought to be a major driver of pathology through hypercitrullination and destabilization of myelin. As such, inhibition of citrullination has been suggested as a therapeutic strategy for MS. Here, in contrast, we show that citrullination by peptidylarginine deiminase 2 (PAD2) contributes to normal oligodendrocyte differentiation, myelination, and motor function. We identify several targets for PAD2, including myelin and chromatin-related proteins, implicating PAD2 in epigenomic regulation. Accordingly, we observe that PAD2 inhibition and its knockdown affect chromatin accessibility and prevent the upregulation of oligodendrocyte differentiation genes. Moreover, mice lacking PAD2 display motor dysfunction and a decreased number of myelinated axons in the corpus callosum. We conclude that citrullination contributes to proper oligodendrocyte lineage progression and myelination.
Keywords: PAD2; Padi2; SILAC; chromatin accessibility; citrullination; deamination; multiple sclerosis; myelin; oligodendrocytes; proteomics.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Assohou-Luty C., Raijmakers R., Benckhuijsen W.E., Stammen-Vogelzangs J., de Ru A., van Veelen P.A., Franken K.L., Drijfhout J.W., Pruijn G.J. The human peptidylarginine deiminases type 2 and type 4 have distinct substrate specificities. Biochim. Biophys. Acta. 2014;1844:829–836. - PubMed
-
- Beniac D.R., Wood D.D., Palaniyar N., Ottensmeyer F.P., Moscarello M.A., Harauz G. Cryoelectron microscopy of protein-lipid complexes of human myelin basic protein charge isomers differing in degree of citrullination. J. Struct. Biol. 2000;129:80–95. - PubMed
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

