Study of chromatin remodeling genes implicates SMARCA4 as a putative player in oncogenesis in neuroblastoma
- PMID: 31018240
- PMCID: PMC6771805
- DOI: 10.1002/ijc.32361
Study of chromatin remodeling genes implicates SMARCA4 as a putative player in oncogenesis in neuroblastoma
Abstract
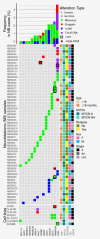
In neuroblastoma (NB), genetic alterations in chromatin remodeling (CRGs) and epigenetic modifier genes (EMGs) have been described. We sought to determine their frequency and clinical impact. Whole exome (WES)/whole genome sequencing (WGS) data and targeted sequencing (TSCA®) of exonic regions of 33 CRGs/EMGs were analyzed in tumor samples from 283 NB patients, with constitutional material available for 55 patients. The frequency of CRG/EMG variations in NB cases was then compared to the Genome Aggregation Database (gnomAD). The sequencing revealed SNVs/small InDels or focal CNAs of CRGs/EMGs in 20% (56/283) of all cases, occurring at a somatic level in 4 (7.2%), at a germline level in 12 (22%) cases, whereas for the remaining cases, only tumor material could be analyzed. The most frequently altered genes were ATRX (5%), SMARCA4 (2.5%), MLL3 (2.5%) and ARID1B (2.5%). Double events (SNVs/small InDels/CNAs associated with LOH) were observed in SMARCA4 (n = 3), ATRX (n = 1) and PBRM1 (n = 1). Among the 60 variations, 24 (8.4%) targeted domains of functional importance for chromatin remodeling or highly conserved domains but of unknown function. Variations in SMARCA4 and ATRX occurred more frequently in the NB as compared to the gnomAD control cohort (OR = 4.49, 95%CI: 1.63-9.97, p = 0.038; OR 3.44, 95%CI: 1.46-6.91, p = 0.043, respectively). Cases with CRG/EMG variations showed a poorer overall survival compared to cases without variations. Genetic variations of CRGs/EMGs with likely functional impact were observed in 8.4% (24/283) of NB. Our case-control approach suggests a role of SMARCA4 as a player of NB oncogenesis.
Keywords: SMARCA4; SWI/SNF; chromatin remodeling complex; epigenetic modifier; neuroblastoma.
© 2019 The Authors. International Journal of Cancer published by John Wiley & Sons Ltd on behalf of UICC.
Figures



References
-
- Masliah‐Planchon J, Bièche I, Guinebretière J‐M, et al. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol Mech Dis 2015;10:145–71. - PubMed
-
- Versteege I, Sévenet N, Lange J, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998;394:203–6. - PubMed
-
- Matthay KK, Maris JM, Schleiermacher G, et al. Neuroblastoma. Nat Rev Dis Primer 2016;2:16078. - PubMed
-
- Molenaar JJ, Koster J, Zwijnenburg DA, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012;483:589–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

