Equine Influenza Virus in Asia: Phylogeographic Pattern and Molecular Features Reveal Circulation of an Autochthonous Lineage
- PMID: 31019053
- PMCID: PMC6580976
- DOI: 10.1128/JVI.00116-19
Equine Influenza Virus in Asia: Phylogeographic Pattern and Molecular Features Reveal Circulation of an Autochthonous Lineage
Abstract
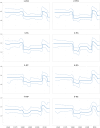
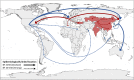
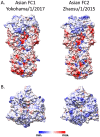
Equine influenza virus (EIV) causes severe acute respiratory disease in horses. Currently, the strains belonging to the H3N8 subtype are divided into two clades, Florida clade 1 (FC1) and Florida clade 2 (FC2), which emerged in 2002. Both FC1 and FC2 clades were reported in Asian and Middle East countries in the last decade. In this study, we described the evolution, epidemiology, and molecular characteristic of the EIV lineages, with focus on those detected in Asia from 2007 to 2017. The full genome phylogeny showed that FC1 and FC2 constituted separate and divergent lineages, without evidence of reassortment between the clades. While FC1 evolved as a single lineage, FC2 showed a divergent event around 2004 giving rise to two well-supported and coexisting sublineages, European and Asian. Furthermore, two different spread patterns of EIV in Asian countries were identified. The FC1 outbreaks were caused by independent introductions of EIV from the Americas, with the Asian isolates genetically similar to the contemporary American lineages. On the other hand, the FC2 strains detected in Asian mainland countries conformed to an autochthonous monophyletic group with a common ancestor dated in 2006 and showed evidence of an endemic circulation in a local host. Characteristic aminoacidic signature patterns were detected in all viral proteins in both Asian-FC1 and FC2 populations. Several changes were located at the top of the HA1 protein, inside or near antigenic sites. Further studies are needed to assess the potential impact of these antigenic changes in vaccination programs.IMPORTANCE The complex and continuous antigenic evolution of equine influenza viruses (EIVs) remains a major hurdle for vaccine development and the design of effective immunization programs. The present study provides a comprehensive analysis showing the EIV evolutionary dynamics, including the spread and circulation within the Asian continent and its relationship to global EIV populations over a 10-year period. Moreover, we provide a better understanding of EIV molecular evolution in Asian countries and its consequences on the antigenicity. The study underscores the association between the global horse movement and the circulation of EIV in this region. Understanding EIV evolution is imperative in order to mitigate the risk of outbreaks affecting the horse industry and to help with the selection of the viral strains to be included in the formulation of future vaccines.
Keywords: Asia; H3N8; H7N7; equine influenza; evolution; influenza; signature pattern; vaccine.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
Emergence of equine influenza virus H3Nx Florida clade 2 in Arabian racehorses in Egypt.Virol J. 2022 Nov 12;19(1):185. doi: 10.1186/s12985-022-01917-9. Virol J. 2022. PMID: 36371185 Free PMC article.
-
Continuing evolution of equine influenza virus in Central Asia, 2007-2012.Arch Virol. 2014 Sep;159(9):2321-7. doi: 10.1007/s00705-014-2078-3. Epub 2014 Apr 20. Arch Virol. 2014. PMID: 24748052
-
Multiple introductions of equine influenza virus into the United Kingdom resulted in widespread outbreaks and lineage replacement.PLoS Pathog. 2025 Jun 9;21(6):e1013227. doi: 10.1371/journal.ppat.1013227. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40489557 Free PMC article.
-
Equine Influenza Virus and Vaccines.Viruses. 2021 Aug 20;13(8):1657. doi: 10.3390/v13081657. Viruses. 2021. PMID: 34452521 Free PMC article. Review.
-
A Brief Introduction to Equine Influenza and Equine Influenza Viruses.Methods Mol Biol. 2020;2123:355-360. doi: 10.1007/978-1-0716-0346-8_26. Methods Mol Biol. 2020. PMID: 32170701 Review.
Cited by
-
Equine Influenza.Cold Spring Harb Perspect Med. 2022 Jan 4;12(1):a038331. doi: 10.1101/cshperspect.a038331. Cold Spring Harb Perspect Med. 2022. PMID: 32152243 Free PMC article. Review.
-
Long-term adaptation following influenza A virus host shifts results in increased within-host viral fitness due to higher replication rates, broader dissemination within the respiratory epithelium and reduced tissue damage.PLoS Pathog. 2021 Dec 17;17(12):e1010174. doi: 10.1371/journal.ppat.1010174. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34919598 Free PMC article.
-
A Cold Case of Equine Influenza Disentangled with Nanopore Sequencing.Animals (Basel). 2023 Mar 24;13(7):1153. doi: 10.3390/ani13071153. Animals (Basel). 2023. PMID: 37048408 Free PMC article.
-
Global Spread of the B5 Subgenotype EV-A71 and the Phylogeographical Analysis of Chinese Migration Events.Front Cell Infect Microbiol. 2020 Sep 25;10:475. doi: 10.3389/fcimb.2020.00475. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33102246 Free PMC article.
-
Equine Mx1 Restricts Influenza A Virus Replication by Targeting at Distinct Site of its Nucleoprotein.Viruses. 2019 Dec 2;11(12):1114. doi: 10.3390/v11121114. Viruses. 2019. PMID: 31810278 Free PMC article.
References
-
- Landolt GA, Townsend HGG, Lunn DP. 2014. Equine influenza infection, p 141–151. In Sellon DC, Long M (ed), Equine infectious diseases 2nd ed Saunders Elsevier, St. Louis, MO.
-
- Wright PF, Neumann G, Kawaoka Y. 2013. Ortomyxoviruses, p 1186–12443. In Knipe D, Howley P (ed), Fields virology 6th ed Wolters Kluwer-Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Ito M, Nagai M, Hayakawa Y, Komae H, Murakami N, Yotsuya S, Asakura S, Sakoda Y, Kida H. 2008. Genetic analyses of an H3N8 influenza virus isolate, causative strain of the outbreak of equine influenza at the Kanazawa racecourse in Japan in 2007. J Vet Med Sci 70:899–906. doi:10.1292/jvms.70.899. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

