Comparative Analysis of African and Asian Lineage-Derived Zika Virus Strains Reveals Differences in Activation of and Sensitivity to Antiviral Innate Immunity
- PMID: 31019057
- PMCID: PMC6580957
- DOI: 10.1128/JVI.00640-19
Comparative Analysis of African and Asian Lineage-Derived Zika Virus Strains Reveals Differences in Activation of and Sensitivity to Antiviral Innate Immunity
Abstract
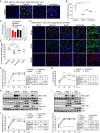
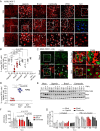
In recent years, Asian lineage Zika virus (ZIKV) strains emerged to cause pandemic outbreaks associated with a high rate of congenital ZIKV syndrome (CZVS). The reasons for the enhanced spread and severe disease caused by newly emerging strains are not fully understood. Here we compared viral sequences, viral replication, and innate immune signaling induction of three different ZIKV strains derived from African and Asian lineages and West Nile virus, another flavivirus. We found pronounced differences in activation of innate immune signaling and inhibition of viral replication across ZIKV strains. The newly emerged Asian ZIKV strain Brazil Fortaleza 2015, which is associated with a higher rate of neurodevelopmental disorders like microcephaly, induced much weaker and delayed innate immune signaling in infected cells. However, superinfection studies to assess control of innate immune signaling induced by Sendai virus argue against an active block of IRF3 activation by the Brazilian strain of ZIKV and rather suggest an evasion of detection by host cell pattern recognition receptors. Compared to the Asian strain FSS13025 isolated in Cambodia, both ZIKV Uganda MR766 and ZIKV Brazil Fortaleza appear less sensitive to the interferon-induced antiviral response. ZIKV infection studies of cells lacking the different RIG-I-like receptors identified RIG-I as the major cytosolic pattern recognition receptor for detection of ZIKV.IMPORTANCE Zika Virus (ZIKV), discovered in 1947, is divided into African and Asian lineages. Pandemic outbreaks caused by currently emerging Asian lineage strains are accompanied by high rates of neurological disorders and exemplify the global health burden associated with this virus. Here we compared virological and innate immunological aspects of two ZIKV strains from the Asian lineage, an emerging Brazilian strain and a less-pathogenic Cambodian strain, and the prototypic African lineage ZIKV strain from Uganda. Compared to the replication of other ZIKV strains, the replication of ZIKV Brazil was less sensitive to the antiviral actions of interferon (IFN), while infection with this strain induced weaker and delayed innate immune responses in vitro Our data suggest that ZIKV Brazil directs a passive strategy of innate immune evasion that is reminiscent of a stealth virus. Such strain-specific properties likely contribute to differential pathogenesis and should be taken into consideration when choosing virus strains for future molecular studies.
Keywords: RIG-I-like receptors; Zika virus; flavivirus; innate immunity.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- PAHO/WHO. 25 August 2017. Zika–epidemiological update. PAHO/WHO, Washington, DC: https://www.paho.org/hq/index.php?option=com_docman&task=doc_download&gi.... Accessed 4 June 2018.
-
- Martines RB, Bhatnagar J, Keating MK, Silva-Flannery L, Muehlenbachs A, Gary J, Goldsmith C, Hale G, Ritter J, Rollin D, Shieh WJ, Luz KG, Ramos AM, Davi HP, Kleber de Oliveria W, Lanciotti R, Lambert A, Zaki S. 2016. Notes from the field: evidence of Zika virus infection in brain and placental tissues from two congenitally infected newborns and two fetal losses–Brazil, 2015. MMWR Morb Mortal Wkly Rep 65:159–160. doi: 10.15585/mmwr.mm6506e1. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

