Human skin long noncoding RNA WAKMAR1 regulates wound healing by enhancing keratinocyte migration
- PMID: 31019085
- PMCID: PMC6511036
- DOI: 10.1073/pnas.1814097116
Human skin long noncoding RNA WAKMAR1 regulates wound healing by enhancing keratinocyte migration
Abstract
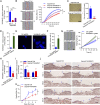
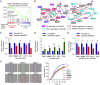
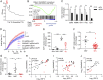
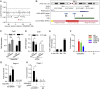
An increasing number of studies reveal the importance of long noncoding RNAs (lncRNAs) in gene expression control underlying many physiological and pathological processes. However, their role in skin wound healing remains poorly understood. Our study focused on a skin-specific lncRNA, LOC105372576, whose expression was increased during physiological wound healing. In human nonhealing wounds, however, its level was significantly lower compared with normal wounds under reepithelialization. We characterized LOC105372576 as a nuclear-localized, RNAPII-transcribed, and polyadenylated lncRNA. In keratinocytes, its expression was induced by TGF-β signaling. Knockdown of LOC105372576 and activation of its endogenous transcription, respectively, reduced and increased the motility of keratinocytes and reepithelialization of human ex vivo skin wounds. Therefore, LOC105372576 was termed "wound and keratinocyte migration-associated lncRNA 1" (WAKMAR1). Further study revealed that WAKMAR1 regulated a network of protein-coding genes important for cell migration, most of which were under the control of transcription factor E2F1. Mechanistically, WAKMAR1 enhanced E2F1 expression by interfering with E2F1 promoter methylation through the sequestration of DNA methyltransferases. Collectively, we have identified a lncRNA important for keratinocyte migration, whose deficiency may be involved in the pathogenesis of chronic wounds.
Keywords: keratinocyte migration; long noncoding RNA; wound healing.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Reinke JM, Sorg H. Wound repair and regeneration. Eur Surg Res. 2012;49:35–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

