Genome-wide analysis of canine oral malignant melanoma metastasis-associated gene expression
- PMID: 31019223
- PMCID: PMC6482147
- DOI: 10.1038/s41598-019-42839-x
Genome-wide analysis of canine oral malignant melanoma metastasis-associated gene expression
Abstract
Oral malignant melanoma (OMM) is the most common canine melanocytic neoplasm. Overlap between the somatic mutation profiles of canine OMM and human mucosal melanomas suggest a shared UV-independent molecular aetiology. In common with human mucosal melanomas, most canine OMM metastasise. There is no reliable means of predicting canine OMM metastasis, and systemic therapies for metastatic disease are largely palliative. Herein, we employed exon microarrays for comparative expression profiling of FFPE biopsies of 18 primary canine OMM that metastasised and 10 primary OMM that did not metastasise. Genes displaying metastasis-associated expression may be targets for anti-metastasis treatments, and biomarkers of OMM metastasis. Reduced expression of CXCL12 in the metastasising OMMs implies that the CXCR4/CXCL12 axis may be involved in OMM metastasis. Increased expression of APOBEC3A in the metastasising OMMs may indicate APOBEC3A-induced double-strand DNA breaks and pro-metastatic hypermutation. DNA double strand breakage triggers the DNA damage response network and two Fanconi anaemia DNA repair pathway members showed elevated expression in the metastasising OMMs. Cross-validation was employed to test a Linear Discriminant Analysis classifier based upon the RT-qPCR-measured expression levels of CXCL12, APOBEC3A and RPL29. Classification accuracies of 94% (metastasising OMMs) and 86% (non-metastasising OMMs) were estimated.
Conflict of interest statement
The authors declare no competing interests.
Figures
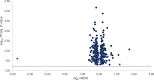
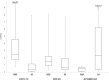
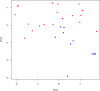
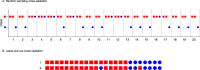
References
-
- Todoroff RJ, Brodey RS. Oral and pharyngeal neoplasia in the dog: a retrospective survey of 361 cases. J. Am. Vet. Med. Assoc. 1979;175:567–571. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

