Ensuring expression of four core cardiogenic transcription factors enhances cardiac reprogramming
- PMID: 31019236
- PMCID: PMC6482135
- DOI: 10.1038/s41598-019-42945-w
Ensuring expression of four core cardiogenic transcription factors enhances cardiac reprogramming
Abstract
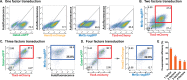
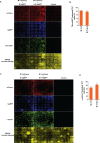
Previous studies have shown that forced expression of core cardiogenic transcription factors can directly reprogram fibroblasts to induced cardiomyocyte-like cells (iCMs). This cardiac reprogramming approach suggests a potential strategy for cardiomyocyte regeneration. However, a major challenge of this approach remains the low conversion rate. Here, we showed that ensuring expression of four cardiogenic transcription factors (i.e. Gata4 (G), Hand2 (H), Mef2c (M), and Tbx5 (T)) in individual fibroblasts is an initial bottleneck for cardiac reprogramming. Following co-transduction of three or four retroviral vectors encoding individual cardiogenic transcription factors, only a minor subpopulation of cells indeed expressed all three (GMT) or four (GHMT) factors. By selectively analyzing subpopulations of cells expressing various combinations of reprogramming factors, we found that co-expression of GMT in individual fibroblasts is sufficient to induce sarcomeric proteins. However, only a small fraction of those cells expressing GMT were able to develop organized sarcomeric structures and contractility. In contrast, ensuring expression of GHMT markedly enhanced the development of contractile cardiac structures and functions in fibroblasts, although its incremental effect on sarcomeric protein induction was relatively small. Our findings provide new insights into the mechanistic basis of inefficient cardiac reprogramming and can help to devise efficient reprogramming strategies.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Stoichiometric optimization of Gata4, Hand2, Mef2c, and Tbx5 expression for contractile cardiomyocyte reprogramming.Sci Rep. 2019 Oct 18;9(1):14970. doi: 10.1038/s41598-019-51536-8. Sci Rep. 2019. PMID: 31628386 Free PMC article.
-
Single-Construct Polycistronic Doxycycline-Inducible Vectors Improve Direct Cardiac Reprogramming and Can Be Used to Identify the Critical Timing of Transgene Expression.Int J Mol Sci. 2017 Aug 19;18(8):1805. doi: 10.3390/ijms18081805. Int J Mol Sci. 2017. PMID: 28825623 Free PMC article.
-
Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors.Development. 2014 Nov;141(22):4267-78. doi: 10.1242/dev.114025. Epub 2014 Oct 24. Development. 2014. PMID: 25344074 Free PMC article.
-
Direct Cardiac Reprogramming: A Novel Approach for Heart Regeneration.Int J Mol Sci. 2018 Sep 5;19(9):2629. doi: 10.3390/ijms19092629. Int J Mol Sci. 2018. PMID: 30189626 Free PMC article. Review.
-
Direct Cardiac Reprogramming for Cardiovascular Regeneration and Differentiation.Keio J Med. 2020 Sep 25;69(3):49-58. doi: 10.2302/kjm.2019-0008-OA. Epub 2020 Jan 9. Keio J Med. 2020. PMID: 31915320 Review.
Cited by
-
The Future of Direct Cardiac Reprogramming: Any GMT Cocktail Variety?Int J Mol Sci. 2020 Oct 26;21(21):7950. doi: 10.3390/ijms21217950. Int J Mol Sci. 2020. PMID: 33114756 Free PMC article. Review.
-
Fibroblast fate determination during cardiac reprogramming by remodeling of actin filaments.Stem Cell Reports. 2022 Jul 12;17(7):1604-1619. doi: 10.1016/j.stemcr.2022.05.004. Epub 2022 Jun 9. Stem Cell Reports. 2022. PMID: 35688153 Free PMC article.
-
Chamber-Specific Protein Expression during Direct Cardiac Reprogramming.Cells. 2021 Jun 16;10(6):1513. doi: 10.3390/cells10061513. Cells. 2021. PMID: 34208439 Free PMC article.
-
A review of protocols for human iPSC culture, cardiac differentiation, subtype-specification, maturation, and direct reprogramming.STAR Protoc. 2022 Aug 18;3(3):101560. doi: 10.1016/j.xpro.2022.101560. eCollection 2022 Sep 16. STAR Protoc. 2022. PMID: 36035804 Free PMC article. Review.
-
Integrase deficient lentiviral vector: prospects for safe clinical applications.PeerJ. 2022 Aug 12;10:e13704. doi: 10.7717/peerj.13704. eCollection 2022. PeerJ. 2022. PMID: 35979475 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

