Tropomyosin concentration but not formin nucleators mDia1 and mDia3 determines the level of tropomyosin incorporation into actin filaments
- PMID: 31019238
- PMCID: PMC6482184
- DOI: 10.1038/s41598-019-42977-2
Tropomyosin concentration but not formin nucleators mDia1 and mDia3 determines the level of tropomyosin incorporation into actin filaments
Abstract
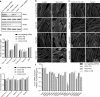
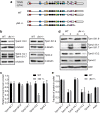
The majority of actin filaments in human cells exist as a co-polymer with tropomyosin, which determines the functionality of actin filaments in an isoform dependent manner. Tropomyosin isoforms are sorted to different actin filament populations and in yeast this process is determined by formins, however it remains unclear what process determines tropomyosin isoform sorting in mammalian cells. We have tested the roles of two major formin nucleators, mDia1 and mDia3, in the recruitment of specific tropomyosin isoforms in mammals. Despite observing poorer cell-cell attachments in mDia1 and mDia3 KD cells and an actin bundle organisation defect with mDia1 knock down; depletion of mDia1 and mDia3 individually and concurrently did not result in any significant impact on tropomyosin recruitment to actin filaments, as observed via immunofluorescence and measured via biochemical assays. Conversely, in the presence of excess Tpm3.1, the absolute amount of Tpm3.1-containing actin filaments is not fixed by actin filament nucleators but rather depends on the cell concentration of Tpm3.1. We conclude that mDia1 and mDia3 are not essential for tropomyosin recruitment and that tropomyosin incorporation into actin filaments is concentration dependent.
Conflict of interest statement
P.W.G. and E.C.H.are directors and shareholders of TroBio Therapeutics, a company that is commercializing anti-tropomyosin drugs for the treatment of cancer and their labs receive funding from TroBio to evaluate anti-tropomyosin drug candidates. J.C.M.M., N.S.B., J.L.G.N., S.S.T. and M.B. declare no financial competing interests. All authors declare no non-financial competing interests.
Figures



Similar articles
-
Tropomyosin isoforms define distinct microfilament populations with different drug susceptibility.Eur J Cell Biol. 2008 Sep;87(8-9):709-20. doi: 10.1016/j.ejcb.2008.03.004. Epub 2008 May 9. Eur J Cell Biol. 2008. PMID: 18472182
-
Tropomyosin - master regulator of actin filament function in the cytoskeleton.J Cell Sci. 2015 Aug 15;128(16):2965-74. doi: 10.1242/jcs.172502. Epub 2015 Aug 3. J Cell Sci. 2015. PMID: 26240174 Review.
-
G-actin regulates rapid induction of actin nucleation by mDia1 to restore cellular actin polymers.J Cell Sci. 2008 Oct 15;121(Pt 20):3403-12. doi: 10.1242/jcs.030940. Epub 2008 Sep 30. J Cell Sci. 2008. PMID: 18827014
-
Recruitment Kinetics of Tropomyosin Tpm3.1 to Actin Filament Bundles in the Cytoskeleton Is Independent of Actin Filament Kinetics.PLoS One. 2016 Dec 15;11(12):e0168203. doi: 10.1371/journal.pone.0168203. eCollection 2016. PLoS One. 2016. PMID: 27977753 Free PMC article.
-
Tropomyosin as a Regulator of Actin Dynamics.Int Rev Cell Mol Biol. 2015;318:255-91. doi: 10.1016/bs.ircmb.2015.06.002. Epub 2015 Jul 7. Int Rev Cell Mol Biol. 2015. PMID: 26315888 Review.
Cited by
-
Caldesmon controls stress fiber force-balance through dynamic cross-linking of myosin II and actin-tropomyosin filaments.Nat Commun. 2022 Oct 13;13(1):6032. doi: 10.1038/s41467-022-33688-w. Nat Commun. 2022. PMID: 36229430 Free PMC article.
-
Diversity from similarity: cellular strategies for assigning particular identities to actin filaments and networks.Open Biol. 2020 Sep;10(9):200157. doi: 10.1098/rsob.200157. Epub 2020 Sep 2. Open Biol. 2020. PMID: 32873155 Free PMC article.
-
Actin-tropomyosin distribution in non-muscle cells.J Muscle Res Cell Motil. 2020 Mar;41(1):11-22. doi: 10.1007/s10974-019-09514-0. Epub 2019 May 4. J Muscle Res Cell Motil. 2020. PMID: 31054005 Free PMC article. Review.
-
The Effect of Tropomyosin Mutations on Actin-Tropomyosin Binding: In Search of Lost Time.Biophys J. 2019 Jun 18;116(12):2275-2284. doi: 10.1016/j.bpj.2019.05.009. Epub 2019 May 13. Biophys J. 2019. PMID: 31130236 Free PMC article.
-
Functional redundancy and formin-independent localization of tropomyosin isoforms in Saccharomyces cerevisiae.bioRxiv [Preprint]. 2024 Dec 11:2024.04.04.587703. doi: 10.1101/2024.04.04.587703. bioRxiv. 2024. PMID: 38617342 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

