Propidium iodide staining underestimates viability of adherent bacterial cells
- PMID: 31019274
- PMCID: PMC6482146
- DOI: 10.1038/s41598-019-42906-3
Propidium iodide staining underestimates viability of adherent bacterial cells
Abstract
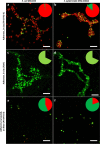
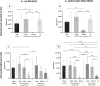
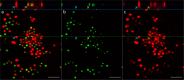
Combining membrane impermeable DNA-binding stain propidium iodide (PI) with membrane-permeable DNA-binding counterstains is a widely used approach for bacterial viability staining. In this paper we show that PI staining of adherent cells in biofilms may significantly underestimate bacterial viability due to the presence of extracellular nucleic acids (eNA). We demonstrate that gram-positive Staphylococcus epidermidis and gram-negative Escherichia coli 24-hour initial biofilms on glass consist of 76 and 96% PI-positive red cells in situ, respectively, even though 68% the cells of either species in these aggregates are metabolically active. Furthermore, 82% of E. coli and 89% S. epidermidis are cultivable after harvesting. Confocal laser scanning microscopy (CLSM) revealed that this false dead layer of red cells is due to a subpopulation of double-stained cells that have green interiors under red coating layer which hints at eNA being stained outside intact membranes. Therefore, viability staining results of adherent cells should always be validated by an alternative method for estimating viability, preferably by cultivation.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- User Manual: LIVE/DEAD BacLight Bacterial Viability Kits. (2004).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

