Self-organization and symmetry breaking in intestinal organoid development
- PMID: 31019299
- PMCID: PMC6544541
- DOI: 10.1038/s41586-019-1146-y
Self-organization and symmetry breaking in intestinal organoid development
Abstract
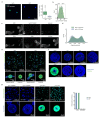
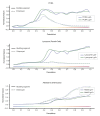
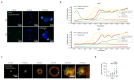
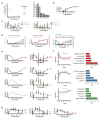
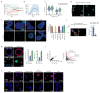
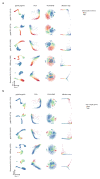
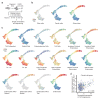
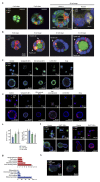
Intestinal organoids are complex three-dimensional structures that mimic the cell-type composition and tissue organization of the intestine by recapitulating the self-organizing ability of cell populations derived from a single intestinal stem cell. Crucial in this process is a first symmetry-breaking event, in which only a fraction of identical cells in a symmetrical sphere differentiate into Paneth cells, which generate the stem-cell niche and lead to asymmetric structures such as the crypts and villi. Here we combine single-cell quantitative genomic and imaging approaches to characterize the development of intestinal organoids from single cells. We show that their development follows a regeneration process that is driven by transient activation of the transcriptional regulator YAP1. Cell-to-cell variability in YAP1, emerging in symmetrical spheres, initiates Notch and DLL1 activation, and drives the symmetry-breaking event and formation of the first Paneth cell. Our findings reveal how single cells exposed to a uniform growth-promoting environment have the intrinsic ability to generate emergent, self-organized behaviour that results in the formation of complex multicellular asymmetric structures.
Conflict of interest statement
Figures





Comment in
-
Yap in regeneration and symmetry breaking.Nat Cell Biol. 2019 Jun;21(6):665-667. doi: 10.1038/s41556-019-0334-1. Nat Cell Biol. 2019. PMID: 31086262 No abstract available.
References
-
- Sasai Y. Cytosystems dynamics in self-organization of tissue architecture. Nature. 2013;493:318–326. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

