Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport
- PMID: 31019304
- PMCID: PMC6750207
- DOI: 10.1038/s41586-019-1135-1
Serotonin transporter-ibogaine complexes illuminate mechanisms of inhibition and transport
Abstract
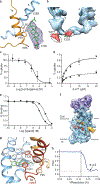
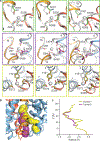
The serotonin transporter (SERT) regulates neurotransmitter homeostasis through the sodium- and chloride-dependent recycling of serotonin into presynaptic neurons1-3. Major depression and anxiety disorders are treated using selective serotonin reuptake inhibitors-small molecules that competitively block substrate binding and thereby prolong neurotransmitter action2,4. The dopamine and noradrenaline transporters, together with SERT, are members of the neurotransmitter sodium symporter (NSS) family. The transport activities of NSSs can be inhibited or modulated by cocaine and amphetamines2,3, and genetic variants of NSSs are associated with several neuropsychiatric disorders including attention deficit hyperactivity disorder, autism and bipolar disorder2,5. Studies of bacterial NSS homologues-including LeuT-have shown how their transmembrane helices (TMs) undergo conformational changes during the transport cycle, exposing a central binding site to either side of the membrane1,6-12. However, the conformational changes associated with transport in NSSs remain unknown. To elucidate structure-based mechanisms for transport in SERT we investigated its complexes with ibogaine, a hallucinogenic natural product with psychoactive and anti-addictive properties13,14. Notably, ibogaine is a non-competitive inhibitor of transport but displays competitive binding towards selective serotonin reuptake inhibitors15,16. Here we report cryo-electron microscopy structures of SERT-ibogaine complexes captured in outward-open, occluded and inward-open conformations. Ibogaine binds to the central binding site, and closure of the extracellular gate largely involves movements of TMs 1b and 6a. Opening of the intracellular gate involves a hinge-like movement of TM1a and the partial unwinding of TM5, which together create a permeation pathway that enables substrate and ion diffusion to the cytoplasm. These structures define the structural rearrangements that occur from the outward-open to inward-open conformations, and provide insight into the mechanism of neurotransmitter transport and ibogaine inhibition.
Conflict of interest statement
The authors declare no competing interests.
Figures










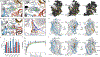
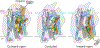
Similar articles
-
Structure-based discovery of conformationally selective inhibitors of the serotonin transporter.Cell. 2023 May 11;186(10):2160-2175.e17. doi: 10.1016/j.cell.2023.04.010. Epub 2023 May 2. Cell. 2023. PMID: 37137306 Free PMC article.
-
Control of serotonin transporter phosphorylation by conformational state.Proc Natl Acad Sci U S A. 2016 May 17;113(20):E2776-83. doi: 10.1073/pnas.1603282113. Epub 2016 May 2. Proc Natl Acad Sci U S A. 2016. PMID: 27140629 Free PMC article.
-
The mechanistic basis for noncompetitive ibogaine inhibition of serotonin and dopamine transporters.J Biol Chem. 2012 May 25;287(22):18524-34. doi: 10.1074/jbc.M112.343681. Epub 2012 Mar 26. J Biol Chem. 2012. PMID: 22451652 Free PMC article.
-
Substrate and inhibitor binding to the serotonin transporter: Insights from computational, crystallographic, and functional studies.Neuropharmacology. 2019 Dec 15;161:107548. doi: 10.1016/j.neuropharm.2019.02.030. Epub 2019 Feb 23. Neuropharmacology. 2019. PMID: 30807752 Review.
-
Emerging structure-function relationships defining monoamine NSS transporter substrate and ligand affinity.Biochem Pharmacol. 2010 Apr 15;79(8):1083-91. doi: 10.1016/j.bcp.2009.11.019. Epub 2009 Nov 30. Biochem Pharmacol. 2010. PMID: 19954741 Review.
Cited by
-
APACE: AlphaFold2 and advanced computing as a service for accelerated discovery in biophysics.Proc Natl Acad Sci U S A. 2024 Jul 2;121(27):e2311888121. doi: 10.1073/pnas.2311888121. Epub 2024 Jun 24. Proc Natl Acad Sci U S A. 2024. PMID: 38913887 Free PMC article.
-
Structures and membrane interactions of native serotonin transporter in complexes with psychostimulants.Proc Natl Acad Sci U S A. 2023 Jul 18;120(29):e2304602120. doi: 10.1073/pnas.2304602120. Epub 2023 Jul 12. Proc Natl Acad Sci U S A. 2023. PMID: 37436958 Free PMC article.
-
Gender Differences in Depression: Evidence From Genetics.Front Genet. 2020 Oct 15;11:562316. doi: 10.3389/fgene.2020.562316. eCollection 2020. Front Genet. 2020. PMID: 33193645 Free PMC article. Review.
-
Monoamine signaling and neuroinflammation: mechanistic connections and implications for neuropsychiatric disorders.Front Immunol. 2025 Apr 28;16:1543730. doi: 10.3389/fimmu.2025.1543730. eCollection 2025. Front Immunol. 2025. PMID: 40356905 Free PMC article. Review.
-
An Overview of Cell-Based Assay Platforms for the Solute Carrier Family of Transporters.Front Pharmacol. 2021 Aug 10;12:722889. doi: 10.3389/fphar.2021.722889. eCollection 2021. Front Pharmacol. 2021. PMID: 34447313 Free PMC article. Review.
References
-
- Kristensen AS et al. SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev 63, 585–640, (2011). - PubMed
-
- Gether U, Andersen PH, Larsson OM & Schousboe A Neurotransmitter transporters: molecular function of important drug targets. Trends Pharmacol Sci 27, 375–383, (2006). - PubMed
-
- Lin Z & Madras BK Human genetics and pharmacology of neurotransmitter transporters. Handb Exp Pharmacol, 327–371, (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

