Prevalence and Diversity of Antibiotic Resistance Genes in Swedish Aquatic Environments Impacted by Household and Hospital Wastewater
- PMID: 31019498
- PMCID: PMC6458280
- DOI: 10.3389/fmicb.2019.00688
Prevalence and Diversity of Antibiotic Resistance Genes in Swedish Aquatic Environments Impacted by Household and Hospital Wastewater
Abstract
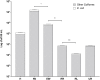
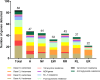
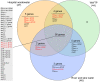
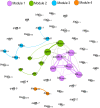
Antibiotic-resistant Enterobacteriaceae and non-lactose fermenting Gram-negative bacteria are a major cause of nosocomial infections. Antibiotic misuse has fueled the worldwide spread of resistant bacteria and the genes responsible for antibiotic resistance (ARGs). There is evidence that ARGs are ubiquitous in non-clinical environments, especially those affected by anthropogenic activity. However, the emergence and primary sources of ARGs in the environment of countries with strict regulations for antibiotics usage are not fully explored. The aim of the present study was to evaluate the repertoire of ARGs of culturable Gram-negative bacteria from directionally connected sites from the hospital to the wastewater treatment plant (WWTP), and downstream aquatic environments in central Sweden. The ARGs were detected from genomic DNA isolated from a population of selectively cultured coliform and Gram-negative bacteria using qPCR. The results show that hospital wastewater was a reservoir of several class B β-lactamase genes such as bla IMP-1 , bla IMP-2, and bla OXA-23, however, most of these genes were not observed in downstream locations. Moreover, β-lactamase genes such as bla OXA-48, bla CTX-M-8, and bla SFC-1, bla V IM-1, and bla V IM-13 were detected in downstream river water but not in the WWTP. The results indicate that the WWTP and hospital wastewaters were reservoirs of most ARGs and contribute to the diversity of ARGs in associated natural environments. However, this study suggests that other factors may also have minor contributions to the prevalence and diversity of ARGs in natural environments.
Keywords: VIM-1; antimicrobial resistance gene co-occurrence; carbapenemase; enterobacteriaceae; extended-spectrum beta-lactamase; surface water; urban wastewater.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

