High-Throughput Phenotyping Enabled Genetic Dissection of Crop Lodging in Wheat
- PMID: 31019521
- PMCID: PMC6459080
- DOI: 10.3389/fpls.2019.00394
High-Throughput Phenotyping Enabled Genetic Dissection of Crop Lodging in Wheat
Abstract
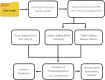
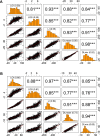
Novel high-throughput phenotyping (HTP) approaches are needed to advance the understanding of genotype-to-phenotype and accelerate plant breeding. The first generation of HTP has examined simple spectral reflectance traits from images and sensors but is limited in advancing our understanding of crop development and architecture. Lodging is a complex trait that significantly impacts yield and quality in many crops including wheat. Conventional visual assessment methods for lodging are time-consuming, relatively low-throughput, and subjective, limiting phenotyping accuracy and population sizes in breeding and genetics studies. Here, we demonstrate the considerable power of unmanned aerial systems (UAS) or drone-based phenotyping as a high-throughput alternative to visual assessments for the complex phenological trait of lodging, which significantly impacts yield and quality in many crops including wheat. We tested and validated quantitative assessment of lodging on 2,640 wheat breeding plots over the course of 2 years using differential digital elevation models from UAS. High correlations of digital measures of lodging to visual estimates and equivalent broad-sense heritability demonstrate this approach is amenable for reproducible assessment of lodging in large breeding nurseries. Using these high-throughput measures to assess the underlying genetic architecture of lodging in wheat, we applied genome-wide association analysis and identified a key genomic region on chromosome 2A, consistent across digital and visual scores of lodging. However, these associations accounted for a very minor portion of the total phenotypic variance. We therefore investigated whole genome prediction models and found high prediction accuracies across populations and environments. This adequately accounted for the highly polygenic genetic architecture of numerous small effect loci, consistent with the previously described complex genetic architecture of lodging in wheat. Our study provides a proof-of-concept application of UAS-based phenomics that is scalable to tens-of-thousands of plots in breeding and genetic studies as will be needed to uncover the genetic factors and increase the rate of gain for complex traits in crop breeding.
Keywords: GWAS; Triticum aestivum; UAV/UAS; genomic selection; high-throughput phenotyping; lodging; unmanned aerial systems; wheat breeding.
Figures



References
-
- Bates D., Maechler M., Bolker B., Walker S. (2014). lme4: linear mixed-effects models using Eigen and S4. R Package Version 1 1–23.
-
- Benaglia T., Chauveau D., Hunter D. R., Young D. S. (2009). mixtools: an R package for analyzing finite mixture models. J. Stat. Softw. 32 1–29. 10.18637/jss.v032.i06 - DOI
-
- Bendig J., Bolten A., Bennertz S., Broscheit J., Eichfuss S., Bareth G. (2014). Estimating biomass of barley using crop surface models (CSMs) Derived from UAV-Based RGB Imaging. Remote Sens. 6 10395–10412. 10.3390/rs61110395 - DOI
-
- Berry P. M., Berry S. T. (2015). Understanding the genetic control of lodging-associated plant characters in winter wheat (Triticum aestivum L.). Euphytica 205 671–689. 10.1007/s10681-015-1387-2 - DOI
Associated data
LinkOut - more resources
Full Text Sources

