Recent advances in understanding the molecular genetic basis of mitochondrial disease
- PMID: 31021000
- PMCID: PMC7041634
- DOI: 10.1002/jimd.12104
Recent advances in understanding the molecular genetic basis of mitochondrial disease
Abstract
Mitochondrial disease is hugely diverse with respect to associated clinical presentations and underlying genetic causes, with pathogenic variants in over 300 disease genes currently described. Approximately half of these have been discovered in the last decade due to the increasingly widespread application of next generation sequencing technologies, in particular unbiased, whole exome-and latterly, whole genome sequencing. These technologies allow more genetic data to be collected from patients with mitochondrial disorders, continually improving the diagnostic success rate in a clinical setting. Despite these significant advances, some patients still remain without a definitive genetic diagnosis. Large datasets containing many variants of unknown significance have become a major challenge with next generation sequencing strategies and these require significant functional validation to confirm pathogenicity. This interface between diagnostics and research is critical in continuing to expand the list of known pathogenic variants and concomitantly enhance our knowledge of mitochondrial biology. The increasing use of whole exome sequencing, whole genome sequencing and other "omics" techniques such as transcriptomics and proteomics will generate even more data and allow further interrogation and validation of genetic causes, including those outside of coding regions. This will improve diagnostic yields still further and emphasizes the integral role that functional assessment of variant causality plays in this process-the overarching focus of this review.
Keywords: diagnosis; mitochondrial disease; molecular mechanisms; next generation sequencing.
© 2019 The Authors. Journal of Inherited Metabolic Disease published by John Wiley & Sons Ltd on behalf of SSIEM.
Conflict of interest statement
Author contributions
K.T., J.J.C.: Review concept, drafting and critical revision of the manuscript, and preparation of tables and figures. R.I.C.G., F.M.R., A.P., E.L.B., C.L.A., M.O.: Drafting and critical revision of the manuscript. R.M., R.W.T.: Review concept, drafting and critical revision of the manuscript.
Figures

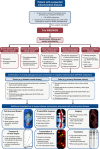
Similar articles
-
Advancing genomic approaches to the molecular diagnosis of mitochondrial disease.Essays Biochem. 2018 Jul 20;62(3):399-408. doi: 10.1042/EBC20170110. Print 2018 Jul 20. Essays Biochem. 2018. PMID: 29950319 Review.
-
Enhanced mitochondrial genome analysis: bioinformatic and long-read sequencing advances and their diagnostic implications.Expert Rev Mol Diagn. 2023 Jul-Dec;23(9):797-814. doi: 10.1080/14737159.2023.2241365. Epub 2023 Aug 29. Expert Rev Mol Diagn. 2023. PMID: 37642407 Review.
-
Next-generation sequencing for mitochondrial disorders.Br J Pharmacol. 2014 Apr;171(8):1837-53. doi: 10.1111/bph.12469. Br J Pharmacol. 2014. PMID: 24138576 Free PMC article. Review.
-
Diagnosis of mitochondrial disorders by concomitant next-generation sequencing of the exome and mitochondrial genome.Genomics. 2013 Sep;102(3):148-56. doi: 10.1016/j.ygeno.2013.04.013. Epub 2013 Apr 28. Genomics. 2013. PMID: 23631824 Free PMC article.
-
The switch in the diagnosis of mitochondrial diseases from the classical 'function first' to the NGS-based 'genetics first' diagnostic era.J Mother Child. 2020 Oct 2;24(2):47-52. doi: 10.34763/jmotherandchild.20202402si.2005.000008. J Mother Child. 2020. PMID: 33179603 Free PMC article. Review.
Cited by
-
Prescription drugs and mitochondrial metabolism.Biosci Rep. 2022 Apr 29;42(4):BSR20211813. doi: 10.1042/BSR20211813. Biosci Rep. 2022. PMID: 35315490 Free PMC article. Review.
-
Neurological Phenotypes in Mouse Models of Mitochondrial Disease and Relevance to Human Neuropathology.Int J Mol Sci. 2023 Jun 2;24(11):9698. doi: 10.3390/ijms24119698. Int J Mol Sci. 2023. PMID: 37298649 Free PMC article. Review.
-
The molecular pathology of pathogenic mitochondrial tRNA variants.FEBS Lett. 2021 Apr;595(8):1003-1024. doi: 10.1002/1873-3468.14049. Epub 2021 Feb 12. FEBS Lett. 2021. PMID: 33513266 Free PMC article. Review.
-
Next-generation sequencing technology in the diagnosis of mitochondrial disorders.Int J Health Sci (Qassim). 2021 Jan-Feb;15(1):1-2. Int J Health Sci (Qassim). 2021. PMID: 33456435 Free PMC article. No abstract available.
-
Mitochondrial diseases: expanding the diagnosis in the era of genetic testing.J Transl Genet Genom. 2020;4:384-428. doi: 10.20517/jtgg.2020.40. Epub 2020 Sep 29. J Transl Genet Genom. 2020. PMID: 33426505 Free PMC article.
References
-
- Rahman S, Thorburn D. Nuclear gene‐encoded Leigh syndrome overview. In: GeneReviews® , ed. Seattle, WA: University of Washington; 2015.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

