Application of a targeted-enrichment methodology for full-genome sequencing of Dengue 1-4, Chikungunya and Zika viruses directly from patient samples
- PMID: 31022183
- PMCID: PMC6504110
- DOI: 10.1371/journal.pntd.0007184
Application of a targeted-enrichment methodology for full-genome sequencing of Dengue 1-4, Chikungunya and Zika viruses directly from patient samples
Abstract
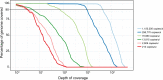
The frequency of epidemics caused by Dengue viruses 1-4, Zika virus and Chikungunya viruses have been on an upward trend in recent years driven primarily by uncontrolled urbanization, mobility of human populations and geographical spread of their shared vectors, Aedes aegypti and Aedes albopictus. Infections by these viruses present with similar clinical manifestations making them challenging to diagnose; this is especially difficult in regions of the world hyperendemic for these viruses. In this study, we present a targeted-enrichment methodology to simultaneously sequence the complete viral genomes for each of these viruses directly from clinical samples. Additionally, we have also developed a customized computational tool (BaitMaker) to design these enrichment baits. This methodology is robust in its ability to capture diverse sequences and is amenable to large-scale epidemiological studies. We have applied this methodology to two large cohorts: a febrile study based in Colombo, Sri Lanka taken during the 2009-2015 dengue epidemic (n = 170) and another taken during the 2016 outbreak of Zika virus in Singapore (n = 162). Results from these studies indicate that we were able to cover an average of 97.04% ± 0.67% of the full viral genome from samples in these cohorts. We also show detection of one DENV3/ZIKV co-infected patient where we recovered full genomes for both viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

