Antibiotic Discovery: Where Have We Come from, Where Do We Go?
- PMID: 31022923
- PMCID: PMC6627412
- DOI: 10.3390/antibiotics8020045
Antibiotic Discovery: Where Have We Come from, Where Do We Go?
Abstract
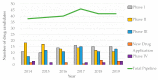
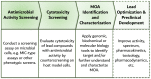
Given the increase in antibiotic-resistant bacteria, alongside the alarmingly low rate of newly approved antibiotics for clinical usage, we are on the verge of not having effective treatments for many common infectious diseases. Historically, antibiotic discovery has been crucial in outpacing resistance and success is closely related to systematic procedures-platforms-that have catalyzed the antibiotic golden age, namely the Waksman platform, followed by the platforms of semi-synthesis and fully synthetic antibiotics. Said platforms resulted in the major antibiotic classes: aminoglycosides, amphenicols, ansamycins, beta-lactams, lipopeptides, diaminopyrimidines, fosfomycins, imidazoles, macrolides, oxazolidinones, streptogramins, polymyxins, sulphonamides, glycopeptides, quinolones and tetracyclines. During the genomics era came the target-based platform, mostly considered a failure due to limitations in translating drugs to the clinic. Therefore, cell-based platforms were re-instituted, and are still of the utmost importance in the fight against infectious diseases. Although the antibiotic pipeline is still lackluster, especially of new classes and novel mechanisms of action, in the post-genomic era, there is an increasingly large set of information available on microbial metabolism. The translation of such knowledge into novel platforms will hopefully result in the discovery of new and better therapeutics, which can sway the war on infectious diseases back in our favor.
Keywords: antibiotic discovery platforms; drug screening; fully synthetic antibiotics; genomics; lipidomics; metabolomics; metagenomics; proteomics; semi-synthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Saga T., Yamaguchi K. History of antimicrobial agents and resistant. Jpn. Med. Assoc. J. 2009;137:103–108.
-
- WHO Report on Global Surveillance of Epidemic-prone Infectious Diseases—Introduction. [(accessed on 12 October 2018)]; Available online: http://www.who.int/csr/resources/publications/introduction/en/index1.html.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

