Identification and characterization of water chestnut Soymovirus-1 (WCSV-1), a novel Soymovirus in water chestnuts (Eleocharis dulcis)
- PMID: 31023231
- PMCID: PMC6482551
- DOI: 10.1186/s12870-019-1761-7
Identification and characterization of water chestnut Soymovirus-1 (WCSV-1), a novel Soymovirus in water chestnuts (Eleocharis dulcis)
Abstract
Background: A disease of unknown etiology in water chestnut plants (Eleocharis dulcis) was reported in China between 2012 and 2014. High throughput sequencing of small RNA (sRNA) combined with bioinformatics, and molecular identification based on PCR detection with virus-specific primers and DNA sequencing is a desirable approach to identify an unknown infectious agent. In this study, we employed this approach to identify viral sequences in water chestnut plants and to explore the molecular interaction of the identified viral pathogen and its natural plant host.
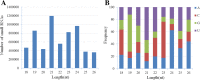
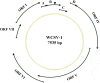
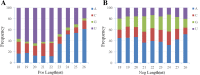
Results: Based on high throughput sequencing of virus-derived small RNAs (vsRNA), we identified the sequence a new-to-science double-strand DNA virus isolated from water chestnut cv. 'Tuanfeng' samples, a widely grown cultivar in Hubei province, China, and analyzed its genomic organization. The complete genomic sequence is 7535 base-pairs in length, and shares 42-52% nucleotide sequence identity with viruses in the Caulimoviridae family. The virus contains nine predicated open reading frames (ORFs) encoding nine hypothetical proteins, with conserved domains characteristic of caulimoviruses. Phylogenetic analyses at the nucleotide and amino acid levels indicated that the virus belongs to the genus Soymovirus. The virus is tentatively named Water chestnut soymovirus-1 (WCSV-1). Phylogenetic analysis of the putative viral polymerase protein suggested that WCSV-1 is distinct to other well established species in the Soymovirus genus. This conclusion was supported by phylogenetic analyses of the amino acid sequences encoded by ORFs I, IV, VI, or VII. The sRNA bioinformatics showed that the majority of the vsRNAs are 22-nt in length with a preference for U at the 5'-terminal nucleotide. The vsRNAs are unevenly distributed over both strands of the entire WCSV-1 circular genome, and are clustered into small defined regions. In addition, we detected WCSV-1 in asymptomatic and symptomatic water chestnut samples collected from different regions of China by using PCR. RNA-seq assays further confirmed the presence of WCSV-1-derived viral RNA in infected plants.
Conclusions: This is the first discovery of a dsDNA virus in the genus Soymovirus infecting water chestnuts. Data presented also add new information towards a better understanding of the co-evolutionary mechanisms between the virus and its natural plant host.
Keywords: Caulimoviridae; Pararetrovirus; RNA sequencing; Soymovirus; Virus-derived small RNA (vsRNA); Water chestnut.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures






References
-
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Ninth report of the international committee on taxonomy of viruses. San Diego: Elsevier Academic Press; 2012.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

