Development and evaluation of a one-step multiplex real-time TaqMan® RT-qPCR assay for the detection and genotyping of equine G3 and G14 rotaviruses in fecal samples
- PMID: 31023319
- PMCID: PMC6482509
- DOI: 10.1186/s12985-019-1149-1
Development and evaluation of a one-step multiplex real-time TaqMan® RT-qPCR assay for the detection and genotyping of equine G3 and G14 rotaviruses in fecal samples
Abstract
Background: Equine rotavirus A (ERVA) is the leading cause of diarrhea in neonatal foals and has a negative impact on equine breeding enterprises worldwide. Among ERVA strains infecting foals, the genotypes G3P[12] and G14P[12] are the most prevalent, while infections by strains with other genomic arrangements are infrequent. The identification of circulating strains of ERVA is critical for diagnostic and surveillance purposes, as well as to understand their molecular epidemiology. Current genotyping methods available for ERVA and rotaviruses affecting other animal species rely on Sanger sequencing and are significantly time-consuming, costly and labor intensive. Here, we developed the first one-step multiplex TaqMan® real-time reverse transcription polymerase chain reaction (RT-qPCR) assay targeting the NSP3 and VP7 genes of ERVA G3 and G14 genotypes for the rapid detection and G-typing directly from fecal specimens.
Methods: A one-step multiplex TaqMan® RT-qPCR assay targeting the NSP3 and VP7 genes of ERVA G3 and G14 genotypes was designed. The analytical sensitivity was assessed using serial dilutions of in vitro transcribed RNA containing the target sequences while the analytical specificity was determined using RNA and DNA derived from a panel of group A rotaviruses along with other equine viruses and bacteria. The clinical performance of this multiplex assay was evaluated using a panel of 177 fecal samples and compared to a VP7-specific standard RT-PCR assay and Sanger sequencing. Limits of detection (LOD), sensitivity, specificity, and agreement were determined.
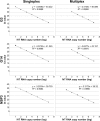
Results: The multiplex G3 and G14 VP7 assays demonstrated high specificity and efficiency, with perfect linearity. A 100-fold difference in their analytical sensitivity was observed when compared to the singleplex assays; however, this difference did not have an impact on the clinical performance. Clinical performance of the multiplex RT-qPCR assay demonstrated that this assay had a high sensitivity/specificity for every target (100% for NSP3, > 90% for G3 VP7 and > 99% for G14 VP7, respectively) and high overall agreement (> 98%) compared to conventional RT-PCR and sequencing.
Conclusions: This new multiplex RT-qPCR assay constitutes a useful, very reliable tool that could significantly aid in the rapid detection and G-typing of ERVA strains circulating in the field.
Keywords: ERVA; Equine rotavirus A; Foal diarrhea; G-typing; G14; G3; One-step multiplex RT-qPCR; Rotavirus A.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Quadruplex Real-Time TaqMan® RT-qPCR Assay for Differentiation of Equine Group A and B Rotaviruses and Identification of Group A G3 and G14 Genotypes.Viruses. 2023 Jul 26;15(8):1626. doi: 10.3390/v15081626. Viruses. 2023. PMID: 37631969 Free PMC article.
-
Detection, molecular characterization and phylogenetic analysis of G3P[12] and G14P[12] equine rotavirus strains co-circulating in central Kentucky.Virus Res. 2018 Aug 15;255:39-54. doi: 10.1016/j.virusres.2018.05.025. Epub 2018 Jun 1. Virus Res. 2018. PMID: 29864502
-
Genetic and antigenic characterization of two diarrhoeicdominant rotavirus A genotypes G3P[12] and G14P[12] circulating in the global equine population.J Gen Virol. 2024 Aug;105(8):002016. doi: 10.1099/jgv.0.002016. J Gen Virol. 2024. PMID: 39163114 Free PMC article.
-
Molecular characterization and analysis of equine rotavirus circulating in Japan from 2003 to 2008.Vet Microbiol. 2011 Aug 26;152(1-2):67-73. doi: 10.1016/j.vetmic.2011.04.016. Epub 2011 Apr 22. Vet Microbiol. 2011. PMID: 21565456
-
Global distribution of group A rotavirus strains in horses: a systematic review.Vaccine. 2013 Nov 19;31(48):5627-33. doi: 10.1016/j.vaccine.2013.08.045. Epub 2013 Aug 28. Vaccine. 2013. PMID: 23994380
Cited by
-
Development and Validation of a Panel of One-Step Four-Plex qPCR/RT-qPCR Assays for Simultaneous Detection of SARS-CoV-2 and Other Pathogens Associated with Canine Infectious Respiratory Disease Complex.Viruses. 2023 Sep 5;15(9):1881. doi: 10.3390/v15091881. Viruses. 2023. PMID: 37766287 Free PMC article.
-
Mouse-Adapted SARS-CoV-2 MA10 Strain Displays Differential Pulmonary Tropism and Accelerated Viral Replication, Neurodissemination, and Pulmonary Host Responses in K18-hACE2 Mice.mSphere. 2023 Feb 21;8(1):e0055822. doi: 10.1128/msphere.00558-22. Epub 2023 Feb 2. mSphere. 2023. PMID: 36728430 Free PMC article.
-
Equine Rotavirus A under the One Health Lens: Potential Impacts on Public Health.Viruses. 2024 Jan 16;16(1):130. doi: 10.3390/v16010130. Viruses. 2024. PMID: 38257830 Free PMC article. Review.
-
Multiplex One-Step RT-qPCR Assays for Simultaneous Detection of SARS-CoV-2 and Other Enteric Viruses of Dogs and Cats.Viruses. 2023 Sep 7;15(9):1890. doi: 10.3390/v15091890. Viruses. 2023. PMID: 37766296 Free PMC article.
-
Equine rotavirus infection.J Equine Sci. 2021 Mar;32(1):1-9. doi: 10.1294/jes.32.1. Epub 2021 Mar 16. J Equine Sci. 2021. PMID: 33776534 Free PMC article. Review.
References
-
- Conner ME, Darlington RW. Rotavirus infection in foals. Am J Vet Res. 1980;41(10):1699–1703. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical