A new widespread subclass of carbonic anhydrase in marine phytoplankton
- PMID: 31024153
- PMCID: PMC6776030
- DOI: 10.1038/s41396-019-0426-8
A new widespread subclass of carbonic anhydrase in marine phytoplankton
Abstract
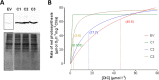
Most aquatic photoautotrophs depend on CO2-concentrating mechanisms (CCMs) to maintain productivity at ambient concentrations of CO2, and carbonic anhydrase (CA) plays a key role in these processes. Here we present different lines of evidence showing that the protein LCIP63, identified in the marine diatom Thalassiosira pseudonana, is a CA. However, sequence analysis showed that it has a low identity with any known CA and therefore belongs to a new subclass that we designate as iota-CA. Moreover, LCIP63 unusually prefers Mn2+ to Zn2+ as a cofactor, which is potentially of ecological relevance since Mn2+ is more abundant than Zn2+ in the ocean. LCIP63 is located in the chloroplast and only expressed at low concentrations of CO2. When overexpressed using biolistic transformation, the rate of photosynthesis at limiting concentrations of dissolved inorganic carbon increased, confirming its role in the CCM. LCIP63 homologs are present in the five other sequenced diatoms and in other algae, bacteria, and archaea. Thus LCIP63 is phylogenetically widespread but overlooked. Analysis of the Tara Oceans database confirmed this and showed that LCIP63 is widely distributed in marine environments and is therefore likely to play an important role in global biogeochemical carbon cycling.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures