ERRα as a Bridge Between Transcription and Function: Role in Liver Metabolism and Disease
- PMID: 31024446
- PMCID: PMC6459935
- DOI: 10.3389/fendo.2019.00206
ERRα as a Bridge Between Transcription and Function: Role in Liver Metabolism and Disease
Abstract
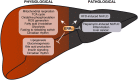
As transcriptional factors, nuclear receptors (NRs) function as major regulators of gene expression. In particular, dysregulation of NR activity has been shown to significantly alter metabolic homeostasis in various contexts leading to metabolic disorders and cancers. The orphan estrogen-related receptor (ERR) subfamily of NRs, comprised of ERRα, ERRβ, and ERRγ, for which a natural ligand has yet to be identified, are known as central regulators of energy metabolism. If AMP-activated protein kinase (AMPK) and mechanistic target of rapamycin (mTOR) can be viewed as sensors of the metabolic needs of a cell and responding acutely via post-translational control of proteins, then the ERRs can be regarded as downstream effectors of metabolism via transcriptional regulation of genes for a long-term and sustained adaptive response. In this review, we will focus on recent findings centered on the transcriptional roles played by ERRα in hepatocytes. Modulation of ERRα activity in both in vitro and in vivo models via genetic or pharmacological manipulation coupled with chromatin-immunoprecipitation (ChIP)-on-chip and ChIP-sequencing (ChIP-seq) studies have been fundamental in delineating the direct roles of ERRα in the control of hepatic gene expression. These studies have identified crucial roles for ERRα in lipid and carbohydrate metabolism as well as in mitochondrial function under both physiological and pathological conditions. The regulation of ERRα expression and activity via ligand-independent modes of action including coregulator binding, post-translational modifications (PTMs) and control of protein stability will be discussed in the context that may serve as valuable tools to modulate ERRα function as new therapeutic avenues for the treatment of hepatic metabolic dysfunction and related diseases.
Keywords: diabetes; high-fat diet; inflammation; liver cancer; metabolism; non-alcoholic fatty liver disease; nuclear receptor.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

