Co-circulation of genetically distinct highly pathogenic avian influenza A clade 2.3.4.4 (H5N6) viruses in wild waterfowl and poultry in Europe and East Asia, 2017-18
- PMID: 31024736
- PMCID: PMC6476160
- DOI: 10.1093/ve/vez004
Co-circulation of genetically distinct highly pathogenic avian influenza A clade 2.3.4.4 (H5N6) viruses in wild waterfowl and poultry in Europe and East Asia, 2017-18
Abstract
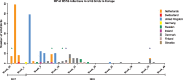
Highly pathogenic avian influenza (HPAI) H5 clade 2.3.4.4 viruses were first introduced into Europe in late 2014 and re-introduced in late 2016, following detections in Asia and Russia. In contrast to the 2014-15 H5N8 wave, there was substantial local virus amplification in wild birds in Europe in 2016-17 and associated wild bird mortality, with evidence for occasional gene exchange with low pathogenic avian influenza (LPAI) viruses. Since December 2017, several European countries have again reported events or outbreaks with HPAI H5N6 reassortant viruses in both wild birds and poultry, respectively. Previous phylogenetic studies have shown that the two earliest incursions of HPAI H5N8 viruses originated in Southeast Asia and subsequently spread to Europe. In contrast, this study indicates that recent HPAI H5N6 viruses evolved from the H5N8 2016-17 viruses during 2017 by reassortment of a European HPAI H5N8 virus and wild host reservoir LPAI viruses. The genetic and phenotypic differences between these outbreaks and the continuing detections of HPAI viruses in Europe are a cause of concern for both animal and human health. The current co-circulation of potentially zoonotic HPAI and LPAI virus strains in Asia warrants the determination of drivers responsible for the global spread of Asian lineage viruses and the potential threat they pose to public health.
Keywords: H5N6; avian influenza; emerging diseases; highly pathogenic avian influenza; phylogeny; virology.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

