Consensus Integrase of a New HIV-1 Genetic Variant CRF63_02A1
- PMID: 31024744
- PMCID: PMC6475865
Consensus Integrase of a New HIV-1 Genetic Variant CRF63_02A1
Abstract
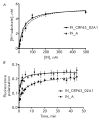
The high genetic variability of the human immunodeficiency virus (HIV-1) leads to a constant emergence of new genetic variants, including the recombinant virus CRF63_02A1, which is widespread in the Siberian Federal District of Russia. We studied HIV-1 CRF63_02A1 integrase (IN_CRF) catalyzing the incorporation of viral DNA into the genome of an infected cell. The consensus sequence was designed, recombinant integrase was obtained, and its DNA-binding and catalytic activities were characterized. The stability of the IN_CRF complex with the DNA substrate did not differ from the complex stability for subtype A and B integrases; however, the rate of complex formation was significantly higher. The rates and efficiencies of 3'-processing and strand transfer reactions catalyzed by IN_CRF were found to be higher, too. Apparently, all these distinctive features of IN_CRF may result from specific amino acid substitutions in its N-terminal domain, which plays an important role in enzyme multimerization and binding to the DNA substrate. It was also found that the drug resistance mutations Q148K/G140S and G118R/E138K significantly reduce the catalytic activity of IN_CRF and its sensitivity to the strand transfer inhibitor raltegravir. Reduction in sensitivity to raltegravir was found to be much stronger in the case of double-mutation Q148K/G140S.
Keywords: CRF63_02A1 genetic variant; drug resistance mutations; human immunodeficiency virus; integrase; strand transfer inhibitor.
Figures





References
-
- https://www.hiv.lanl.gov/content/ sequence/HIV/CRFs/CRFs.html. Foley B., Leitner T., Apetrei C., Hahn B., Mizrachi I., Mullins J., Rambaut A., Wolinsky S., Korber B., Theoretical Biology and Biophysics Group. Los Alamos: Los Alamos National Laboratory. 2013.
-
- Hemelaar J., Trends Mol Med. 2012;13(3):182–192. - PubMed
-
- Diez-Fuertes F., Cabello M., Thompson M.M.. Infect Genet Evol. 2015;33:197–205. - PubMed
LinkOut - more resources
Full Text Sources
