Kinetics of Enzymatic Mercury Methylation at Nanomolar Concentrations Catalyzed by HgcAB
- PMID: 31028026
- PMCID: PMC6581168
- DOI: 10.1128/AEM.00438-19
Kinetics of Enzymatic Mercury Methylation at Nanomolar Concentrations Catalyzed by HgcAB
Abstract
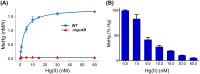
Methylmercury (MeHg) is a potent bioaccumulative neurotoxin that is produced by certain anaerobic bacteria and archaea. Mercury (Hg) methylation has been linked to the gene pair hgcAB, which encodes a membrane-associated corrinoid protein and a ferredoxin. Although microbial Hg methylation has been characterized in vivo, the cellular biochemistry and the specific roles of the gene products HgcA and HgcB in Hg methylation are not well understood. Here, we report the kinetics of Hg methylation in cell lysates of Desulfovibrio desulfuricans ND132 at nanomolar Hg concentrations. The enzymatic Hg methylation mediated by HgcAB is highly oxygen sensitive, irreversible, and follows Michaelis-Menten kinetics, with an apparent Km of 3.2 nM and Vmax of 19.7 fmol · min-1 · mg-1 total protein for the substrate Hg(II). Although the abundance of HgcAB in the cell lysates is extremely low, Hg(II) was quantitatively converted to MeHg at subnanomolar substrate concentrations. Interestingly, increasing thiol/Hg(II) ratios did not impact Hg methylation rates, which suggests that HgcAB-mediated Hg methylation effectively competes with cellular thiols for Hg(II), consistent with the low apparent Km Supplementation of 5-methyltetrahydrofolate or pyruvate did not enhance MeHg production, while both ATP and a nonhydrolyzable ATP analog decreased Hg methylation rates in cell lysates under the experimental conditions. These studies provide insights into the biomolecular processes associated with Hg methylation in anaerobic bacteria.IMPORTANCE The concentration of Hg in the biosphere has increased dramatically over the last century as a result of industrial activities. The microbial conversion of inorganic Hg to MeHg is a global public health concern due to bioaccumulation and biomagnification of MeHg in food webs. Exposure to neurotoxic MeHg through the consumption of fish represents a significant risk to human health and can result in neuropathies and developmental disorders. Anaerobic microbial communities in sediments and periphyton biofilms have been identified as sources of MeHg in aquatic systems, but the associated biomolecular mechanisms are not fully understood. In the present study, we investigate the biochemical mechanisms and kinetics of MeHg formation by HgcAB in sulfate-reducing bacteria. These findings advance our understanding of microbial MeHg production and may help inform strategies to limit the formation of MeHg in the environment.
Keywords: HgcAB; anaerobic bacteria; environmental microbiology; enzyme kinetics; mercury methylation; methylmercury.
Copyright © 2019 American Society for Microbiology.
Figures



References
-
- United Nations Environment Programme. 2013. Global Mercury Assessment 2013: sources, emissions, releases, and environmental transport. UNEP Chemicals Branch, Geneva, Switzerland.
-
- Castoldi AF, Coccini T, Manzo L. 2003. Neurotoxic and molecular effects of methylmercury in humans. Rev Environ Health 18:19–31. - PubMed
-
- Ranchou-Peyruse M, Monperrus M, Bridou R, Duran R, Amouroux D, Salvado JC, Guyoneaud R. 2009. Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers: implications for environmental studies. Geomicrobiol J 26:1–8. doi:10.1080/01490450802599227. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

