Characterisation of class VI TRIM RING domains: linking RING activity to C-terminal domain identity
- PMID: 31028095
- PMCID: PMC6487577
- DOI: 10.26508/lsa.201900295
Characterisation of class VI TRIM RING domains: linking RING activity to C-terminal domain identity
Abstract
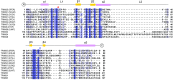
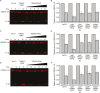
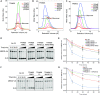
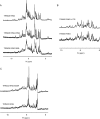
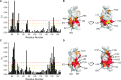
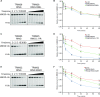
TRIM E3 ubiquitin ligases regulate multiple cellular processes, and their dysfunction is linked to disease. They are characterised by a conserved N-terminal tripartite motif comprising a RING, B-box domains, and a coiled-coil region, with C-terminal domains often mediating substrate recruitment. TRIM proteins are grouped into 11 classes based on C-terminal domain identity. Class VI TRIMs, TRIM24, TRIM33, and TRIM28, have been described as transcriptional regulators, a function linked to their C-terminal plant homeodomain and bromodomain, and independent of their ubiquitination activity. It is unclear whether E3 ligase activity is regulated in family members where the C-terminal domains function independently. Here, we provide a detailed biochemical characterisation of the RING domains of class VI TRIMs and describe the solution structure of the TRIM28 RING. Our study reveals a lack of activity of the isolated RING domains, which may be linked to the absence of self-association. We propose that class VI TRIMs exist in an inactive state and require additional regulatory events to stimulate E3 ligase activity, ensuring that associated chromatin-remodelling factors are not injudiciously degraded.
© 2019 Stevens et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
