Dynamic super-resolution structured illumination imaging in the living brain
- PMID: 31028150
- PMCID: PMC6511017
- DOI: 10.1073/pnas.1819965116
Dynamic super-resolution structured illumination imaging in the living brain
Abstract
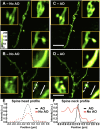
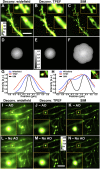
Cells in the brain act as components of extended networks. Therefore, to understand neurobiological processes in a physiological context, it is essential to study them in vivo. Super-resolution microscopy has spatial resolution beyond the diffraction limit, thus promising to provide structural and functional insights that are not accessible with conventional microscopy. However, to apply it to in vivo brain imaging, we must address the challenges of 3D imaging in an optically heterogeneous tissue that is constantly in motion. We optimized image acquisition and reconstruction to combat sample motion and applied adaptive optics to correcting sample-induced optical aberrations in super-resolution structured illumination microscopy (SIM) in vivo. We imaged the brains of live zebrafish larvae and mice and observed the dynamics of dendrites and dendritic spines at nanoscale resolution.
Keywords: adaptive optics; brain imaging; in vivo; super-resolution; synapses.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kandel ER, Schwartz JH, Jessell TM, Siegelbaum S, Hudspeth AJ, editors. Principles of Neural Science. 5th Ed McGraw-Hill Education/Medical; New York: 2012.
-
- Tønnesen J, Nägerl UV. Superresolution imaging for neuroscience. Exp Neurol. 2013;242:33–40. - PubMed
-
- Tønnesen J, Katona G, Rózsa B, Nägerl UV. Spine neck plasticity regulates compartmentalization of synapses. Nat Neurosci. 2014;17:678–685. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

