Ancient Polyploidy and Genome Evolution in Palms
- PMID: 31028709
- PMCID: PMC6535811
- DOI: 10.1093/gbe/evz092
Ancient Polyploidy and Genome Evolution in Palms
Abstract
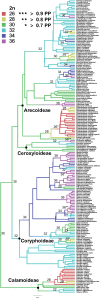
Mechanisms of genome evolution are fundamental to our understanding of adaptation and the generation and maintenance of biodiversity, yet genome dynamics are still poorly characterized in many clades. Strong correlations between variation in genomic attributes and species diversity across the plant tree of life suggest that polyploidy or other mechanisms of genome size change confer selective advantages due to the introduction of genomic novelty. Palms (order Arecales, family Arecaceae) are diverse, widespread, and dominant in tropical ecosystems, yet little is known about genome evolution in this ecologically and economically important clade. Here, we take a phylogenetic comparative approach to investigate palm genome dynamics using genomic and transcriptomic data in combination with a recent, densely sampled, phylogenetic tree. We find conclusive evidence of a paleopolyploid event shared by the ancestor of palms but not with the sister clade, Dasypogonales. We find evidence of incremental chromosome number change in the palms as opposed to one of recurrent polyploidy. We find strong phylogenetic signal in chromosome number, but no signal in genome size, and further no correlation between the two when correcting for phylogenetic relationships. Palms thus add to a growing number of diverse, ecologically successful clades with evidence of whole-genome duplication, sister to a species-poor clade with no evidence of such an event. Disentangling the causes of genome size variation in palms moves us closer to understanding the genomic conditions facilitating adaptive radiation and ecological dominance in an evolutionarily successful, emblematic tropical clade.
Keywords: Arecaceae; Arecales; chromosome; dysploidy; genome duplication; genome size.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Automat Contr. 19(6):716–723.
-
- Asmussen CB, et al. 2006. A new subfamily classification of the palm family (Arecaceae): evidence from plastid DNA phylogeny. Bot J Linn Soc. 151(1):15–38.
-
- Bacon CD, Baker WJ, Simmons MP.. 2012. Miocene dispersal drives island radiations in the palm tribe Trachycarpeae (Arecaceae). Syst Biol. 61(3):426–442. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

